Hi community
Im try to load this data set but the columns name are in bad format. Im put the col_type as date but don't work.
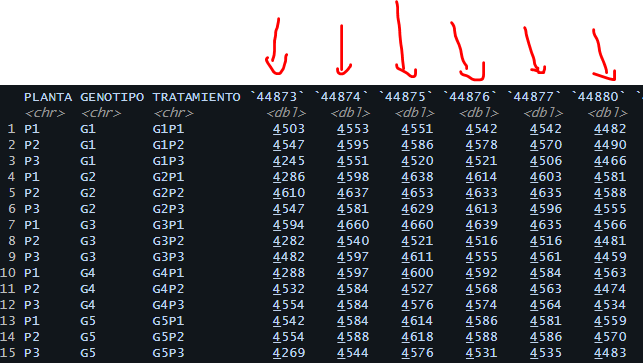
When Im load the data, this is the result:
ACUTIF_R2 <- read_excel("path.xlsx", col_names = T)
# The name of column is a date format (check the link)
data
PLANTA,GENOTIPO,TRATAMIENTO,8-nov-22,9-nov-22,10-nov-22,11-nov-22,12-nov-22,15-nov-22,16-nov-22,17-nov-22,18-nov-22,19-nov-22,21-nov-22,22-nov-22,23-nov-22,24-nov-22,25-nov-22,26-nov-22,28-nov-22
P1,G1,G1P1,4503,4553,4551,4542,4542,4482,4501,45...
Next I'm want make a pivot for get better data for plot and statistics.
library(tidyverse)
ACUTIF_R2 %>% pivot_longer(cols=c('44873', '44874'),
names_to='FECHA',
values_to='PESO')
Thanks for any guide.
R doesn't have the concept of data types for column names, Excell can have different data types for each cell but that is not possible in R. I think the easiest solution would be to manually change the cell format of your column names to "text" in Excell before reading it into R because cleaning it in R afterward is going to be very hacky.
1 Like
Ok, Im understand.
I think you have to use a formula like =TEXTO(D1; "d-mmm-yy"). I'll take a look into an R solution anyways and post it later if no one beat me to the punch.
1 Like
library(tidyverse)
library(readxl)
file_url <- "https://docs.google.com/spreadsheets/d/1RLl8mmZzoC_nyu20Y8iJ3cgSwLlUn2wc/export?format=xlsx"
download.file(file_url, "path.xlsx")
ACUTIF_R2 <- read_excel("path.xlsx", col_names = T)
names(ACUTIF_R2)[str_detect(names(ACUTIF_R2), "\\d{5}")] <- format(as.Date(as.numeric(names(ACUTIF_R2)[str_detect(names(ACUTIF_R2), "\\d{5}")]), origin = "1899-12-30"), "%d-%b-%y")
ACUTIF_R2 %>%
mutate(across(matches("\\d{1,2}-"), as.numeric)) %>%
pivot_longer(cols = matches("\\d{1,2}-"),
names_to='FECHA',
values_to='PESO')
#> Warning: There were 42 warnings in `mutate()`.
#> The first warning was:
#> ℹ In argument: `across(matches("\\d{1,2}-"), as.numeric)`.
#> Caused by warning:
#> ! NAs introducidos por coerción
#> ℹ Run `dplyr::last_dplyr_warnings()` to see the 41 remaining warnings.
#> # A tibble: 780 × 5
#> PLANTA GENOTIPO TRATAMIENTO FECHA PESO
#> <chr> <chr> <chr> <chr> <dbl>
#> 1 P1 G1 G1P1 08-nov-22 4503
#> 2 P1 G1 G1P1 09-nov-22 4553
#> 3 P1 G1 G1P1 10-nov-22 4551
#> 4 P1 G1 G1P1 11-nov-22 4542
#> 5 P1 G1 G1P1 12-nov-22 4542
#> 6 P1 G1 G1P1 15-nov-22 4482
#> 7 P1 G1 G1P1 16-nov-22 4501
#> 8 P1 G1 G1P1 17-nov-22 4509
#> 9 P1 G1 G1P1 18-nov-22 4520
#> 10 P1 G1 G1P1 19-nov-22 4513
#> # … with 770 more rows
Created on 2023-01-31 with reprex v2.0.2
1 Like
Was excellent!
system
February 7, 2023, 8:11pm
7
This topic was automatically closed 7 days after the last reply. New replies are no longer allowed.