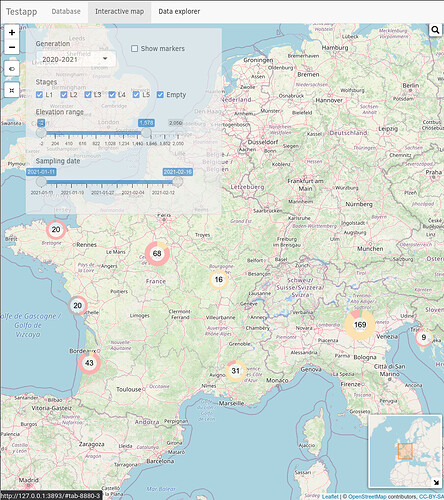
Hi,
I want to plot Leaflet markers corresponding to different levels of the same factor, so this type of clustering is absolutely perfect for me. I successfully used a variation of that in the code below, but I want to use leafletProxy() to avoid redrawing the base map (tiles, position and zoom level) every time an input is altered. Therefore I separated the base map in renderLeaflet() and the data layer (addAwesomeMarkers()) which I put into leafletProxy() in an observer().
Problems
The main problem is addAwesomeMarkers() does not show markers or clusters when I put it in observer(), whereas it does when I include it in renderLeaflet() directly:
in renderLeaflet()` (but of course that beats the point, the full map is redrawn every time inputs are touched):
In observer():
ss-2022-01-16_055642|444x500 (I can only embed one image as a new user)
After some iterations, I realized the problem is specifically with my clusterOptions(): if I remove that option, then the red markers will show with the data layer from the observer(), and the map won't be redrawn all the time. But I need this option to get donut clusters. Any idea why it works outside the observer() and not within it?
Another issue is markers are supposed to be of different colours for every level of the Instar factor, just as in the stackoverflow example, but as you can see they are all red: https://user-images.githubusercontent.com/80409402/149648464-933f1f3d-8656-483f-ab46-d6defb45d70a.png (I can only post two links as a new user :/).
Code
The below code is lengthy, I am sorry. Every time I tried to simplify it to keep only what is relevant, I broke everything and had to debug, so I thought it might be more useful anyway to show the full thing in case my issue is elsewhere. Some things are not working, please disregard them (the "Show markers" is just a placeholder for now, same for the buttons in the third tab).
# Environment and data ####
if (!require("pacman")) install.packages("pacman", repos = "https://pbil.univ-lyon1.fr/CRAN/")
pacman::p_load(leaflet, leaflet.extras, ggplot2, wesanderson, RSQLite, shiny, lubridate, shinydashboard, dplyr, data.table, ggthemes, plyr)
setwd("/path/to/data/")
# Connect to db
db <- dbConnect(drv = RSQLite::SQLite(), dbname = "db/data.db")
# List all tables
tables <- dbListTables(db)
# Exclude sqlite_sequence (contains table information)
tables <- tables[tables != "sqlite_sequence"]
lDataFrames <- vector("list", length=length(tables))
# Create a data.frame for each table
for (i in seq(along = tables)) {
lDataFrames[[i]] <- dbGetQuery(conn = db, statement = paste("SELECT * FROM '", tables[[i]], "'", sep = ""))
}
# Summary of samples ####
# Store raw data table into object rawdata and adjust variable classes
rawdata <- data.table(lDataFrames[[1]])
rawdata$Generation <- as.factor(ifelse(month(rawdata$Date) < 5,
paste0(year(rawdata$Date) - 1,
"-",
year(rawdata$Date)),
paste0(year(rawdata$Date),
"-",
year(rawdata$Date) + 1)))
rawdata$Instar <- factor(rawdata$Instar, levels = c("L1", "L2", "L3", "L4", "L5", "Empty"))
rawdata$Tree <- factor(rawdata$Tree, levels = c("Cedrus deodara", "Pinus brutia", "Pinus halepensis", "Pinus nigra",
"Pinus pinaster", "Pinus pinea", "Pinus radiata", "Pinus sylvestris",
"Pseudotsuga menziesii", "Procession"))
rawdata$Date <- as.Date(rawdata$Date)
rawdata$Region <- as.factor(rawdata$Region)
rawdata$OriginalRegion <- as.factor(rawdata$OriginalRegion)
rawdata$Country <- as.factor(rawdata$Country)
rawdata$n <- as.integer(rawdata$n)
rawdata$Longitude <- as.numeric(rawdata$Longitude)
rawdata$Latitude <- as.numeric(rawdata$Latitude)
rawdata$Elevation <- as.numeric(rawdata$Elevation)
samples <- data.frame(matrix(NA, nrow = length(levels(rawdata$Generation)), ncol = 3))
for (i in 1:nlevels(rawdata$Generation)) {
samples[i,1] <- levels(rawdata$Generation)[i]
samples[i,2] <- sum(subset(rawdata, rawdata$Generation == levels(rawdata$Generation)[i])$n)
samples[i,3] <- nlevels(droplevels(subset(rawdata, rawdata$Generation == levels(rawdata$Generation)[i])$Country))
}
names(samples) <- c("Generation", "Tents", "Countries")
ui <- navbarPage("Testapp ", id="nav", selected = "Interactive map",
tabPanel("Database"),
tabPanel("Interactive map",
div(class="outer",
tags$head(
# Include our custom CSS
includeCSS("styles.css"),
includeScript("gomap.js")
),
# If not using custom CSS, set height of leafletOutput to a number instead of percent
leafletOutput("map", width="100%", height="100%"),
# Shiny versions prior to 0.11 should use class = "modal" instead.
absolutePanel(id = "controls", class = "panel panel-default", fixed = TRUE,
draggable = TRUE, top = 60, left = 55, right = "auto", bottom = "auto",
width = 400, height = 380,
h2(),
fluidRow(
column(6,
selectInput("generation", "Generation",
choices = levels(rawdata$Generation),
selected = max(levels(rawdata$Generation)))
),
column(6,
checkboxInput("markers", "Show markers")
)
),
fluidRow(
column(12,
checkboxGroupInput("instar", "Stages",
choices = levels(rawdata$Instar),
selected = levels(rawdata$Instar),
inline = TRUE)
)
),
fluidRow(
column(12,
sliderInput("elevation", "Elevation range",
min = min(rawdata$Elevation),
max = max(rawdata$Elevation),
value = c(min(rawdata$Elevation), max(rawdata$Elevation)))
)
),
fluidRow(
column(12,
uiOutput("date")
)
)
),
tags$div(id="cite",
'Placeholder'
)
)
),
tabPanel("Data explorer",
fluidRow(
column(3, offset = 0,
style='padding-left: 40px; background: #EEEEEE;',
fluidRow(
selectInput("generation", "Generation",
choices = levels(rawdata$Generation),
selected = max(levels(rawdata$Generation)))
),
fluidRow(
checkboxGroupInput("tree", "Host tree",
choices = levels(rawdata$Tree),
selected = levels(rawdata$Tree),
inline = FALSE)
),
fluidRow(
column(6,
numericInput("minScore", "Min score", min=0, max=100, value=0)
),
column(6,
numericInput("maxScore", "Max score", min=0, max=100, value=100)
)
)
),
column(4,
plotOutput("plot2", height = 340)
),
column(5,
"Sample size per generation",
plotOutput("samples", height = 340)
),
),
hr(),
div(DT::dataTableOutput("tab"),
style = "font-size: 80%;"
),
hr(),
div(
downloadButton("downloadCsv", "Download as CSV"),
verbatimTextOutput("rawtable")
)
),
conditionalPanel("false", icon("crosshair"))
)
server <- function(input, output, session) {
tmpdata <- reactive({
rawdata %>%
filter(Generation == input$generation)
})
samplesbarplot <- reactive({
ggplot(data = samples, aes(x = Generation)) +
geom_bar(aes(y = Tents), alpha = 0.7, fill = wes_palette("Chevalier1")[3], stat = "identity") +
geom_bar(aes(y = Tents), data = subset(samples, samples$Generation == input$generation), alpha = 0.4,
fill = "#B2B2FD", stat = "identity") +
geom_line(aes(y = Countries * 20, group = 1), alpha = 0.3, color = wes_palette("Royal2")[5], lwd = 2) +
geom_point(aes(y = Countries * 20, group = 1), alpha = 1, color = wes_palette("Royal2")[5], size = 4) +
geom_text(aes(label = paste(Tents, "tents"), y = Tents - 50), color = wes_palette("Moonrise2")[1], vjust = 0, size = 4) +
geom_text(aes(label = paste(Countries, "\ncountries"), y = Countries * 20 - 160, size = 3.5),
color = wes_palette("Royal2")[5], vjust = 0, size = 4) +
ylim(c(0, 1000)) +
labs(title = NULL, x = NULL, y = NULL) +
theme_minimal() +
theme(
text = element_text(family = "Arial", color = "#22211d"),
axis.line.y = element_blank(),
panel.grid.major = element_line(color = "#EDEDE9", size = 0.3),
panel.grid.minor = element_blank(),
plot.background = element_rect(fill = "#F8FAFB", color = NA), # other nice vintage colour: #f5f5f2
panel.background = element_rect(fill = "#F8FAFB", color = NA), # other nice vintage colour: #f5f5f2
legend.background = element_rect(fill = "#F8FAFB", color = NA)) # other nice vintage colour: #f5f5f2
})
# Marker colors
colpalette <- c("#E1BBF9", "#B2B2FD", "#B2D8B2", "#FDE3B2", "#FDB2B2", "#BBBBBB")[1:nlevels(rawdata$Instar)]
getColor <- function(x) {colpalette[x$Instar]}
icons <- makeAwesomeIcon(
text = ~substr(Instar, 1, 2),
markerColor = getColor(rawdata)
)
# Javascript for dynamic clusters
jsscript3<- paste0(
"function(cluster) {
const groups = [",paste("'", levels(rawdata$Instar), "'", sep = "", collapse = ","),"];
const colors = {
groups: [", paste("'", colpalette,"'", sep = "",collapse = ","),"],
center: '#ddd',
text: 'black'
};
const markers = cluster.getAllChildMarkers();
const proportions = groups.map(group => markers.filter(marker => marker.options.group === group).length / markers.length);
function sum(arr, first = 0, last) {
return arr.slice(first, last).reduce((total, curr) => total+curr, 0);
}
const cumulativeProportions = proportions.map((val, i, arr) => sum(arr, 0, i+1));
cumulativeProportions.unshift(0);
const width = 1.3*Math.sqrt(markers.length);
const radius = 15+width/2;
const arcs = cumulativeProportions.map((prop, i) => { return {
x : radius*Math.sin(2*Math.PI*prop),
y : -radius*Math.cos(2*Math.PI*prop),
long: proportions[i-1] >.5 ? 1 : 0
}});
const paths = proportions.map((prop, i) => {
if (prop === 0) return '';
else if (prop === 1) return `<circle cx = '0' cy = '0' r = '${radius}' fill = 'none' stroke = '${colors.groups[i]}' stroke-width = '${width}' stroke-alignment = 'center' stroke-linecap = 'butt' />`;
else return `<path d='M ${arcs[i].x} ${arcs[i].y} A ${radius} ${radius} 0 ${arcs[i+1].long} 1 ${arcs[i+1].x} ${arcs[i+1].y}' fill = 'none' stroke = '${colors.groups[i]}' stroke-width = '${width}' stroke-alignment = 'center' stroke-linecap = 'butt' />`
});
return new L.DivIcon({
html: `
<svg width = '200' height = '200' viewBox = '-100 -100 200 200' style = 'width: 200px; height: 200px; position: relative; top: -94px; left: -94px;' >
<circle cx = '0' cy = '0' r = '15' stroke = 'none' fill = '#FFFFFF99' />
<text x = '0' y = '0' dominant-baseline = 'central' text-anchor = 'middle' fill = '${colors.text}' font-size = '15'>${markers.length}</text>
${paths.join('')}
</svg>
`,
className: 'marker-cluster'
});
}")
# Base map
output$map <- renderLeaflet({
leaflet() %>%
addTiles() %>%
setView(6.7250000, 42.366667, zoom = 6) %>%
addMiniMap(
toggleDisplay = TRUE) %>%
addEasyButton(easyButton(
states = list(
easyButtonState(
stateName = "unfrozen-markers",
icon = "ion-toggle",
title = "Freeze clusters regardless\nof zoom level",
onClick = JS("function(btn, map) {
var clusterManager =
map.layerManager.getLayer('cluster', 'donuts');
clusterManager.freezeAtZoom();
btn.state('frozen-markers');
}")
),
easyButtonState(
stateName = "frozen-markers",
icon = "ion-toggle-filled",
title = "Unfreeze clusters",
onClick = JS("function(btn, map) {
var clusterManager =
map.layerManager.getLayer('cluster', 'donuts');
clusterManager.unfreeze();
btn.state('unfrozen-markers');
}")
)
)
)) %>%
addSearchOSM(options = searchOptions(position = "topright",
zoom = 8,
autoCollapse = TRUE,
hideMarkerOnCollapse = TRUE,
minLength = 2)) %>%
addResetMapButton()
# addAwesomeMarkers(data = tmpdata2(), # Code for adding dynamic clusters, but redraws the map on every change
# group = ~Instar, # and marker colors are all the same
# icon = icons,
# clusterOptions = markerClusterOptions(
# iconCreateFunction = JS(jsscript3)),
# clusterId = "donuts")
})
# Observe and leafletProxy to avoid redrawing the map every time markers change, but clusters don't work for no clear reason
observe({
leafletProxy("map", data = tmpdata2()) %>%
clearMarkers() %>%
addAwesomeMarkers(group = ~Instar,
icon = icons,
popup = "test",
label = "label",
#clusterOptions = markerClusterOptions(
# iconCreateFunction = JS(jsscript3)),
clusterId = "donuts")
})
output$samples <- renderPlot(samplesbarplot())
output$tab <- DT::renderDataTable({
df <- tmpdata2() %>%
filter(Generation == input$generation,
Instar %in% input$instar,
Elevation >= input$elevation[1],
Elevation <= input$elevation[2],
Date >= input$date[1],
Date <= input$date[2],
Tree %in% input$tree)
action <- DT::dataTableAjax(session, df, outputId = "tab")
DT::datatable(df, options = list(ajax = list(url = action)), escape = FALSE)
})
output$rawtable <- renderPrint({
df <- tmpdata2() %>%
filter(Generation == input$generation,
Instar %in% input$instar,
Elevation >= input$elevation[1],
Elevation <= input$elevation[2],
Date >= input$date[1],
Date <= input$date[2],
Tree %in% input$tree)
orig <- options(width = 1000)
print(tail(df, 200), row.names = FALSE)
options(orig)
})
output$downloadCsv <- downloadHandler(
filename = function() {"PCLM_excerpt.csv"},
content = function(file) {
df <- tmpdata2() %>%
filter(Generation == input$generation,
Instar %in% input$instar,
Elevation >= input$elevation[1],
Elevation <= input$elevation[2],
Date >= input$date[1],
Date <= input$date[2],
Tree %in% input$tree)
write.csv(df, file)
}
)
output$date <- renderUI({
sliderInput("date", "Sampling date",
min = min(tmpdata()$Date),
max = max(tmpdata()$Date),
value = c(min(tmpdata()$Date), max(tmpdata()$Date)))
})
tmpdata2 <- reactive({
req(input$date)
rawdata %>%
filter(Generation == input$generation,
Instar %in% input$instar,
Elevation >= input$elevation[1],
Elevation <= input$elevation[2],
Date >= input$date[1],
Date <= input$date[2]
)
})
}
shinyApp(ui, server)
These are the problematic bits:
# Base map
output$map <- renderLeaflet({
leaflet() %>%
[…]
addResetMapButton()
# addAwesomeMarkers(data = tmpdata2(), # Code for adding dynamic clusters, but redraws the map on every change
# group = ~Instar, # and marker colors are all the same
# icon = icons,
# clusterOptions = markerClusterOptions(
# iconCreateFunction = JS(jsscript3)),
# clusterId = "donuts")
})
Commenting out the block (and adding a pipe) will show clusters but the base map is refreshed all the time, and markers are all the same color as opposed to the stackoverflow example.
# Observe and leafletProxy to avoid redrawing the map every time markers change, but clusters don't work for no clear reason
observe({
leafletProxy("map", data = tmpdata2()) %>%
clearMarkers() %>%
addAwesomeMarkers(group = ~Instar,
icon = icons,
popup = "test",
label = "label",
#clusterOptions = markerClusterOptions(
# iconCreateFunction = JS(jsscript3)),
clusterId = "donuts")
})
Using that instead will solve the map refreshing issue, but it only works if clusterOptions() is commented out, and the marker color issue is still there.
Am I missing something obvious? Thank you for making it up to the end of this post!