Hello
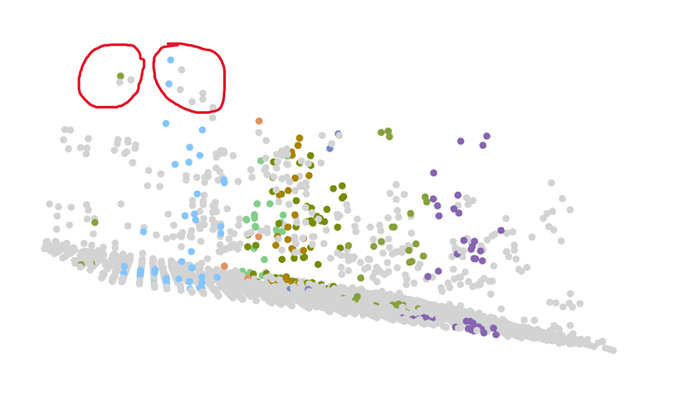
In short, I'm trying to calculate the density of trees over 7m tall, so I want to estimate the crown of each tree in my test plot. To do this, I'm trying to segment each tree over 7m to give to the crown_metrics function. I've tried different approaches with the segment_trees function, using different algorithms (dalponte2016, silva2016, etc), and I've also tried changing different parameters in the algorithms. So far, dalponte2016 has given me the best result, but I'm still not satisfied as some points are not well segmented (I tried to take a screenshot, I'm not sure it's clear enough). As you can see (red circle), some points are not segmented even though they are clearly part of the tree.
The only possible explanation that I'm thinking of is that my point density isn't sufficient (4.6 points/m²).
Or that I'm not using the algorithm correctly.
Here's my code (I didn't put all the attempts with different parameters for dalponte2016):
las <- lidR::readLAS('Data/placette2.las')
las.norm <- lidR::normalize_height(test.placette, algorithm = tin(),na.rm =T, res= 0.3)
las.norm <- filter_poi(las.norm, Z >= 0, Z<50)
chm <- rasterize_canopy(las.norm, res = 0.5, p2r(0.2, na.fill=tin()))
w <- matrix(1,3,3)
chm.smoothed <- raster::focal(chm, w, fun = mean, na.rm =T)
ttops <- locate_trees(las.norm, lmf(ws = 3))
ttops <- ttops[which(ttops$Z >= 7),]
algo <- dalponte2016(chm.smoothed,ttops)
las.segmented <- segment_trees(las.norm, algo)
plot(las, bg = "white", size = 8, color = "treeID")
crowns <- crown_metrics(las.segmented, func = .stdtreemetrics, geom = "convex")
plot(crowns$geometry, col =height.colors(25), main ="Crown with Tree tops")
plot(sf::st_geometry(ttops), add = TRUE, pch = 3)
I wanted my script to be a reproducible example, but I don't really know how to do that with .LAS files.
Best Regards,
Lea