Since your time column is of type character, it is easy to pull out just the time portion of each entry and filter on that or use grepl to detect 12:00:00 in the time column.
kyt <- structure(list(time = c("2022-08-30T12:00:00Z", "2022-08-30T11:50:00Z",
"2022-08-30T11:40:00Z", "2022-08-30T11:30:00Z", "2022-08-30T11:20:00Z",
"2022-08-30T11:10:00Z", "2022-08-30T11:00:00Z", "2022-08-30T10:50:00Z",
"2022-08-30T10:40:00Z", "2022-08-30T10:30:00Z", "2022-08-30T10:20:00Z",
"2022-08-30T10:10:00Z", "2022-08-30T10:00:00Z", "2022-08-30T09:50:00Z",
"2022-08-30T09:40:00Z", "2022-08-30T09:30:00Z", "2022-08-30T09:20:00Z",
"2022-08-30T09:10:00Z", "2022-08-30T09:00:00Z", "2022-08-30T08:50:00Z"
), temp_dry = c(20.9, 21, 20.4, 20.4, 20.7, 20.4, 19.7, 19.7,
20.4, 19.7, 19.5, 19.5, 19.6, 19.4, 19.5, 18.9, 19, 18.2, 18.2,
18.8)), row.names = c(NA, -20L), class = c("tbl_df", "tbl", "data.frame"
))
kyt$TimeOnly <- substr(kyt$time,12, 19)
kyt
#> time temp_dry TimeOnly
#> 1 2022-08-30T12:00:00Z 20.9 12:00:00
#> 2 2022-08-30T11:50:00Z 21.0 11:50:00
#> 3 2022-08-30T11:40:00Z 20.4 11:40:00
#> 4 2022-08-30T11:30:00Z 20.4 11:30:00
#> 5 2022-08-30T11:20:00Z 20.7 11:20:00
#> 6 2022-08-30T11:10:00Z 20.4 11:10:00
#> 7 2022-08-30T11:00:00Z 19.7 11:00:00
#> 8 2022-08-30T10:50:00Z 19.7 10:50:00
#> 9 2022-08-30T10:40:00Z 20.4 10:40:00
#> 10 2022-08-30T10:30:00Z 19.7 10:30:00
#> 11 2022-08-30T10:20:00Z 19.5 10:20:00
#> 12 2022-08-30T10:10:00Z 19.5 10:10:00
#> 13 2022-08-30T10:00:00Z 19.6 10:00:00
#> 14 2022-08-30T09:50:00Z 19.4 09:50:00
#> 15 2022-08-30T09:40:00Z 19.5 09:40:00
#> 16 2022-08-30T09:30:00Z 18.9 09:30:00
#> 17 2022-08-30T09:20:00Z 19.0 09:20:00
#> 18 2022-08-30T09:10:00Z 18.2 09:10:00
#> 19 2022-08-30T09:00:00Z 18.2 09:00:00
#> 20 2022-08-30T08:50:00Z 18.8 08:50:00
library(dplyr)
FilterKYT <- kyt |> filter(TimeOnly == "12:00:00")
FilterKYT
#> # A tibble: 1 × 3
#> time temp_dry TimeOnly
#> <chr> <dbl> <chr>
#> 1 2022-08-30T12:00:00Z 20.9 12:00:00
#Method 2, use grepl
kyt <- structure(list(time = c("2022-08-30T12:00:00Z", "2022-08-30T11:50:00Z",
"2022-08-30T11:40:00Z", "2022-08-30T11:30:00Z", "2022-08-30T11:20:00Z",
"2022-08-30T11:10:00Z", "2022-08-30T11:00:00Z", "2022-08-30T10:50:00Z",
"2022-08-30T10:40:00Z", "2022-08-30T10:30:00Z", "2022-08-30T10:20:00Z",
"2022-08-30T10:10:00Z", "2022-08-30T10:00:00Z", "2022-08-30T09:50:00Z",
"2022-08-30T09:40:00Z", "2022-08-30T09:30:00Z", "2022-08-30T09:20:00Z",
"2022-08-30T09:10:00Z", "2022-08-30T09:00:00Z", "2022-08-30T08:50:00Z"
), temp_dry = c(20.9, 21, 20.4, 20.4, 20.7, 20.4, 19.7, 19.7,
20.4, 19.7, 19.5, 19.5, 19.6, 19.4, 19.5, 18.9, 19, 18.2, 18.2,
18.8)), row.names = c(NA, -20L), class = c("tbl_df", "tbl", "data.frame"
))
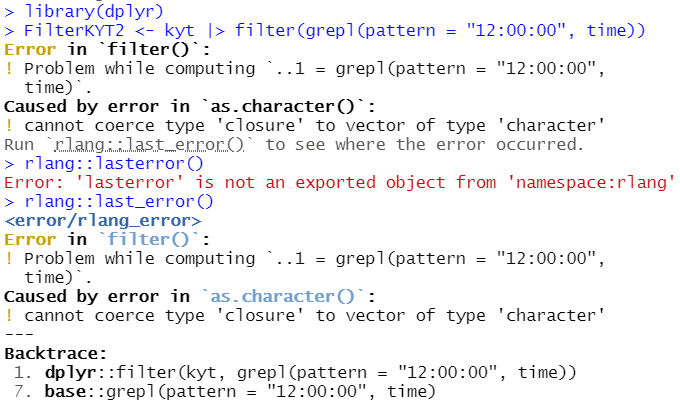
FilterKYT2 <- kyt |> filter(grepl(pattern = "12:00:00", time))
FilterKYT2
#> # A tibble: 1 × 2
#> time temp_dry
#> <chr> <dbl>
#> 1 2022-08-30T12:00:00Z 20.9
Created on 2022-12-09 with reprex v2.0.2
Logical subscripts must match the size of the indexed input.
Input has size 2 but subscript