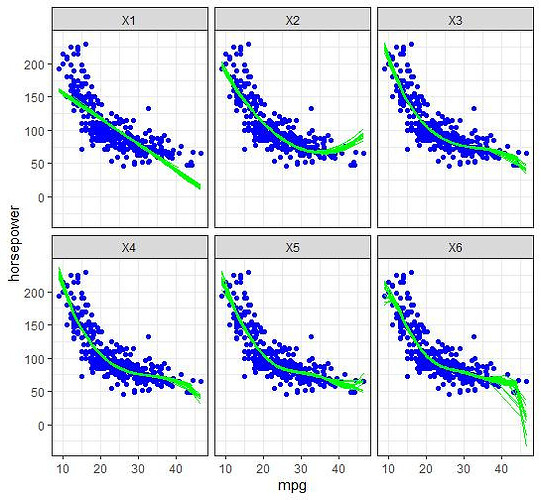
Hi all. I am training data on various regression models (x to x^6), and then using it to predict on a test set. I am having trouble with visualization. By randomly splitting my data 50 times and then training/predicting, ultimately I want to output:
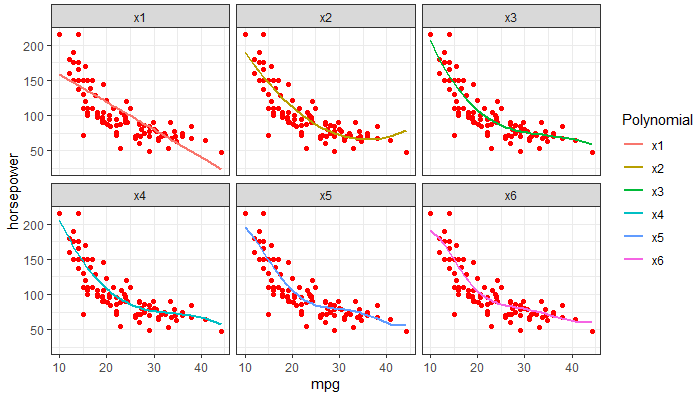
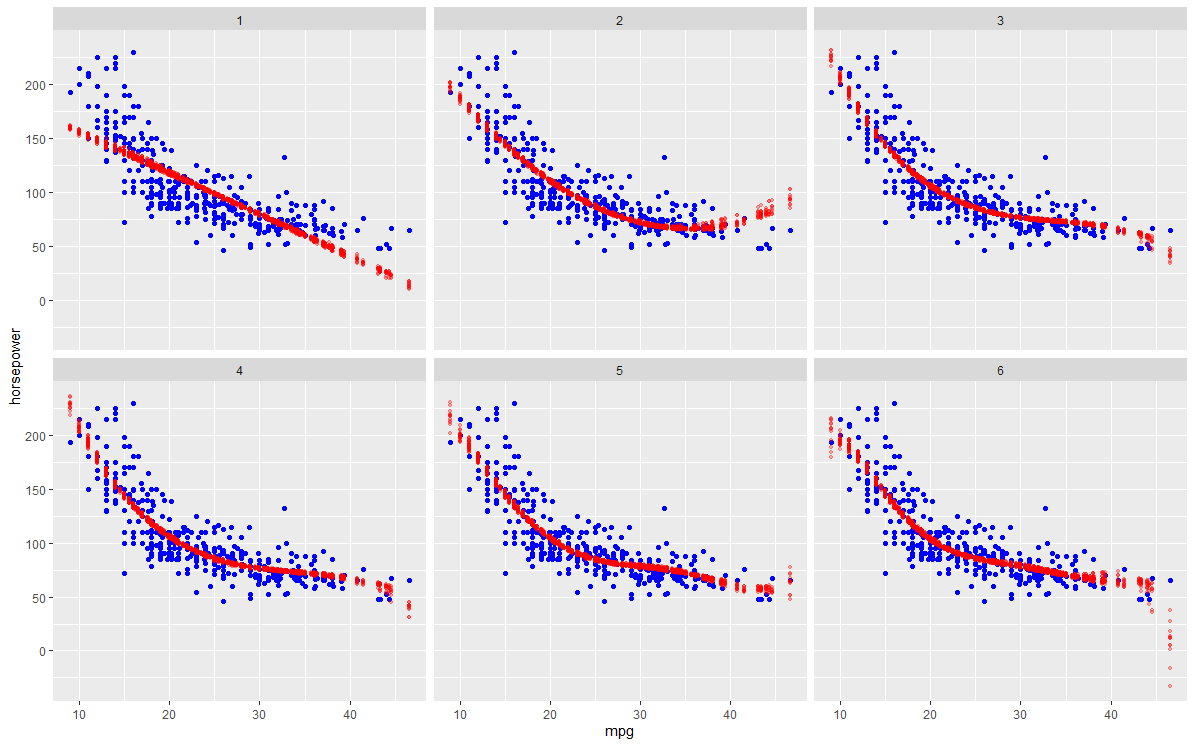
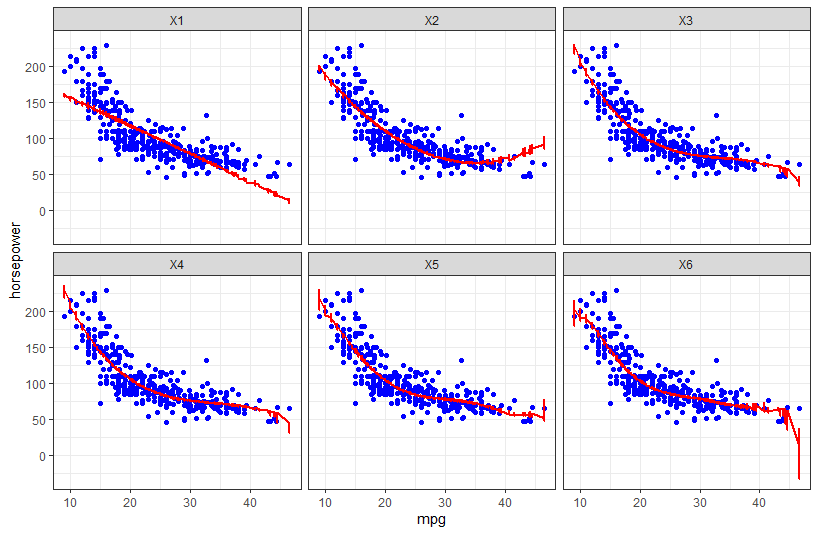
- For each polynomial, the 50 separate prediction lines drawn on the same plot so as to visualize the variance. As for the scatterplot in the background, I assume it should be a scatter of the full dataset?
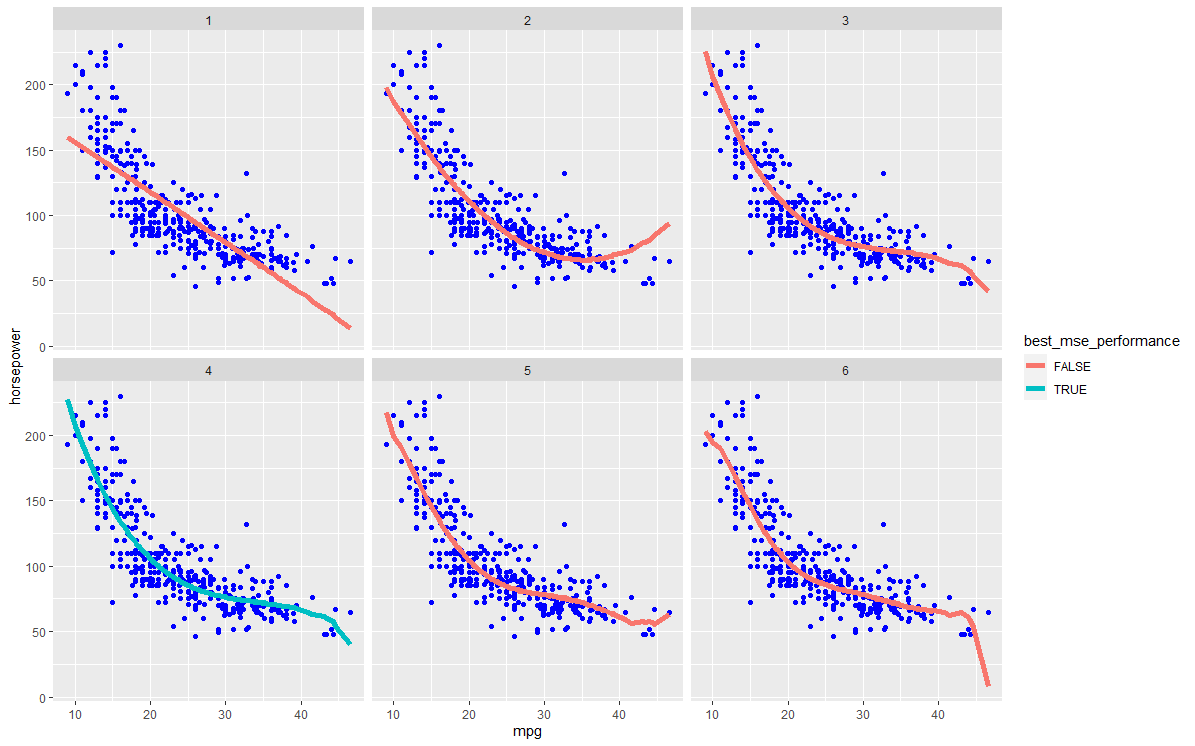
- Highlight the test error of the best model across the 50 iterations. The error lines of other models can be coloured in a lighter colour.
In the code below, I have so far been able to achieve:
(i) Storing the test set mean sq errors (MSEs) for each model across 50 iterations in all.mse.dat
(ii) Plots with a single prediction line for each polynomial (not inside the loop). The lines are against the scatterplot of a single test set (instead of the full dataset).
# use Auto dataset from ISLR
library(ISLR)
str(Auto)
# Create df/vector to store MSEs
# for each of six models across 50 iterations
all.mse.dat <- data.frame(matrix(data = NA,
ncol = 6,
nrow = 50))
mse.test <- vector(mode = "numeric", length = 6)
# Begin the iteration
set.seed(1337)
for (j in 1:50){
# sample random train and test sets (75%/25% split)
rows.test <- sample(1:nrow(Auto),
size = nrow(Auto)*0.25,
replace = FALSE)
train.dat <- Auto[-rows.test, c("mpg", "horsepower") ]
test.dat <- Auto[rows.test, c("mpg", "horsepower") ]
# loop to train and predict
for (i in 1:6){
# train models
model_name <- paste0("train", i,".lm")
assign(model_name, lm(horsepower ~ poly(mpg, i), data = train.dat))
# get predictions
fit_name <- paste0("test", i,".pred")
assign(fit_name, predict(get(paste0("train", i,".lm")), newdata = test.dat))
# get prediction MSE of each model and store them
mse.test[i] <- mean((get(fit_name) - test.dat$horsepower)^2)
all.mse.dat[j, ] <- mse.test
}
} # not sure how to do the below in a loop, or if it is necessary, so I have left the rest outside for now
# Create data.frame for the predictions
test.pred.dat <- data.frame(matrix(data = NA,
ncol = 2+6,
nrow = nrow(test.dat)))
colnames(test.pred.dat) <- c("mpg", "horsepower", paste0("x", 1:6))
# fill the data frame
test.pred.dat$mpg <- test.dat$mpg
test.pred.dat$horsepower <- test.dat$horsepower
for (i in 1:6){
test.pred.dat[i + 2] <- get(paste0("test", i,".pred"))
}
# reshape wide to long for ggplot
require(reshape2)
long.testpred.dat <- reshape2::melt(test.pred.dat,
id.vars = c("mpg", "horsepower"),
measure.vars = c(paste0("x", 1:6)),
variable.name = "Polynomial",
value.name = "y.fit")
# ggplot
require(ggplot2)
ggplot(long.testpred.dat, aes(x = mpg, y = horsepower,
color = Polynomial)) +
geom_point(color = "red") + # need scatter of full Auto dataset instead of just test set
geom_line(aes(y = y.fit), size = 1) +
facet_wrap(~ Polynomial)
I'm also not quite sure if I understand how/what to visualise for 2), in case my wording seems confusing.
Thanks for any help!