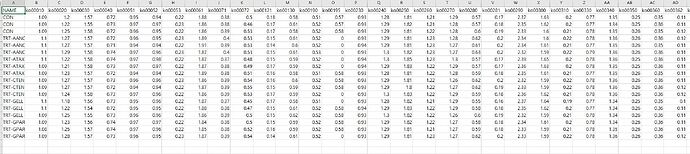
Actually, i am beginner of R software, using nest function is difficult to me.
type or paste code here
library(tidyverse)
library(readxl)
library(reprex)
library(datapasta)
kegg <- read_csv(file = "raw_data/kegg relative.csv")
kegg %>% group_by(NAME) %>% summarise(n = n(), mean = mean(ko00072), sd = sd(ko00072))
kruskal.test(ko00072 ~ NAME, data = kegg)
type or paste code here
kegg <- tibble::tribble(
~NAME, ~ko00010, ~ko00020, ~ko00030, ~ko00040, ~ko00051, ~ko00052, ~ko00053, ~ko00061, ~ko00071,
"CON", 1.09, 1.2, 1.57, 0.72, 0.95, 0.94, 0.22, 1.88, 0.38,
"CON", 1.09, 1.22, 1.55, 0.73, 0.97, 0.97, 0.23, 1.86, 0.38,
"CON", 1.09, 1.25, 1.58, 0.72, 0.96, 0.95, 0.22, 1.86, 0.39,
"TRT-AANC", 1.1, 1.27, 1.57, 0.72, 0.96, 0.95, 0.23, 1.89, 0.4,
"TRT-AANC", 1.1, 1.27, 1.57, 0.71, 0.94, 0.94, 0.22, 1.91, 0.39,
"TRT-AANC", 1.1, 1.29, 1.58, 0.74, 0.97, 0.96, 0.23, 1.87, 0.4,
"TRT-ATAX", 1.1, 1.22, 1.58, 0.74, 0.97, 0.98, 0.22, 1.87, 0.37,
"TRT-ATAX", 1.09, 1.21, 1.58, 0.73, 0.97, 0.97, 0.22, 1.87, 0.38,
"TRT-ATAX", 1.09, 1.23, 1.57, 0.72, 0.94, 0.94, 0.22, 1.89, 0.38,
"TRT-CTEN", 1.09, 1.27, 1.57, 0.73, 0.96, 0.96, 0.23, 1.86, 0.39,
"TRT-CTEN", 1.09, 1.27, 1.57, 0.72, 0.94, 0.94, 0.22, 1.87, 0.39,
"TRT-CTEN", 1.09, 1.24, 1.58, 0.73, 0.97, 0.96, 0.22, 1.86, 0.39,
"TRT-GELL", 1.1, 1.18, 1.56, 0.73, 0.95, 0.96, 0.22, 1.87, 0.37,
"TRT-GELL", 1.1, 1.22, 1.54, 0.72, 0.95, 0.95, 0.22, 1.88, 0.38,
"TRT-GELL", 1.09, 1.25, 1.55, 0.73, 0.96, 0.96, 0.22, 1.86, 0.39,
"TRT-GPAR", 1.09, 1.23, 1.56, 0.74, 0.97, 0.97, 0.22, 1.84, 0.38,
"TRT-GPAR", 1.08, 1.25, 1.57, 0.74, 0.97, 0.96, 0.22, 1.85, 0.38,
"TRT-GPAR", 1.09, 1.28, 1.57, 0.73, 0.96, 0.95, 0.23, 1.87, 0.39
)
head(kegg)
library(tidyverse)
library(readxl)
library(reprex)
library(datapasta)
kegg <- tibble::tribble(
~NAME, ~ko00010, ~ko00020, ~ko00030, ~ko00040, ~ko00051, ~ko00052, ~ko00053, ~ko00061, ~ko00071,
"CON", 1.09, 1.2, 1.57, 0.72, 0.95, 0.94, 0.22, 1.88, 0.38,
"CON", 1.09, 1.22, 1.55, 0.73, 0.97, 0.97, 0.23, 1.86, 0.38,
"CON", 1.09, 1.25, 1.58, 0.72, 0.96, 0.95, 0.22, 1.86, 0.39,
"TRT-AANC", 1.1, 1.27, 1.57, 0.72, 0.96, 0.95, 0.23, 1.89, 0.4,
"TRT-AANC", 1.1, 1.27, 1.57, 0.71, 0.94, 0.94, 0.22, 1.91, 0.39,
"TRT-AANC", 1.1, 1.29, 1.58, 0.74, 0.97, 0.96, 0.23, 1.87, 0.4,
"TRT-ATAX", 1.1, 1.22, 1.58, 0.74, 0.97, 0.98, 0.22, 1.87, 0.37,
"TRT-ATAX", 1.09, 1.21, 1.58, 0.73, 0.97, 0.97, 0.22, 1.87, 0.38,
"TRT-ATAX", 1.09, 1.23, 1.57, 0.72, 0.94, 0.94, 0.22, 1.89, 0.38,
"TRT-CTEN", 1.09, 1.27, 1.57, 0.73, 0.96, 0.96, 0.23, 1.86, 0.39,
"TRT-CTEN", 1.09, 1.27, 1.57, 0.72, 0.94, 0.94, 0.22, 1.87, 0.39,
"TRT-CTEN", 1.09, 1.24, 1.58, 0.73, 0.97, 0.96, 0.22, 1.86, 0.39,
"TRT-GELL", 1.1, 1.18, 1.56, 0.73, 0.95, 0.96, 0.22, 1.87, 0.37,
"TRT-GELL", 1.1, 1.22, 1.54, 0.72, 0.95, 0.95, 0.22, 1.88, 0.38,
"TRT-GELL", 1.09, 1.25, 1.55, 0.73, 0.96, 0.96, 0.22, 1.86, 0.39,
"TRT-GPAR", 1.09, 1.23, 1.56, 0.74, 0.97, 0.97, 0.22, 1.84, 0.38,
"TRT-GPAR", 1.08, 1.25, 1.57, 0.74, 0.97, 0.96, 0.22, 1.85, 0.38,
"TRT-GPAR", 1.09, 1.28, 1.57, 0.73, 0.96, 0.95, 0.23, 1.87, 0.39
)
head(kegg)
#> # A tibble: 6 x 10
#> NAME ko00010 ko00020 ko00030 ko00040 ko00051 ko00052 ko00053 ko00061 ko00071
#>
#> 1 CON 1.09 1.2 1.57 0.72 0.95 0.94 0.22 1.88 0.38
#> 2 CON 1.09 1.22 1.55 0.73 0.97 0.97 0.23 1.86 0.38
#> 3 CON 1.09 1.25 1.58 0.72 0.96 0.95 0.22 1.86 0.39
#> 4 TRT-A~ 1.1 1.27 1.57 0.72 0.96 0.95 0.23 1.89 0.4
#> 5 TRT-A~ 1.1 1.27 1.57 0.71 0.94 0.94 0.22 1.91 0.39
#> 6 TRT-A~ 1.1 1.29 1.58 0.74 0.97 0.96 0.23 1.87 0.4
I just copy bit of my data in CSV file.
Thanks!