One thing I really want to achieve is to assign the data() uploaded by the user to data, so that I can use data directly afterwards without the need for data().
Thank you very much!
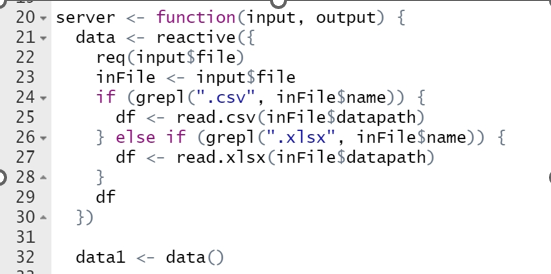
One thing I really want to achieve is to assign the data() uploaded by the user to data, so that I can use data directly afterwards without the need for data().
Thank you very much!

Line 32 illustrates perfectly the problem with using names like data for user-created objects. It's easy to lose track of the context.
data
#> function (..., list = character(), package = NULL, lib.loc = NULL,
#> verbose = getOption("verbose"), envir = .GlobalEnv, overwrite = TRUE)
#> {
#> fileExt <- function(x) {
#> db <- grepl("\\.[^.]+\\.(gz|bz2|xz)$", x)
#> ans <- sub(".*\\.", "", x)
#> ans[db] <- sub(".*\\.([^.]+\\.)(gz|bz2|xz)$", "\\1\\2",
#> x[db])
#> ans
#> }
#> my_read_table <- function(...) {
#> lcc <- Sys.getlocale("LC_COLLATE")
#> on.exit(Sys.setlocale("LC_COLLATE", lcc))
#> Sys.setlocale("LC_COLLATE", "C")
#> read.table(...)
#> }
#> stopifnot(is.character(list))
#> names <- c(as.character(substitute(list(...))[-1L]), list)
#> if (!is.null(package)) {
#> if (!is.character(package))
#> stop("'package' must be a character vector or NULL")
#> }
#> paths <- find.package(package, lib.loc, verbose = verbose)
#> if (is.null(lib.loc))
#> paths <- c(path.package(package, TRUE), if (!length(package)) getwd(),
#> paths)
#> paths <- unique(normalizePath(paths[file.exists(paths)]))
#> paths <- paths[dir.exists(file.path(paths, "data"))]
#> dataExts <- tools:::.make_file_exts("data")
#> if (length(names) == 0L) {
#> db <- matrix(character(), nrow = 0L, ncol = 4L)
#> for (path in paths) {
#> entries <- NULL
#> packageName <- if (file_test("-f", file.path(path,
#> "DESCRIPTION")))
#> basename(path)
#> else "."
#> if (file_test("-f", INDEX <- file.path(path, "Meta",
#> "data.rds"))) {
#> entries <- readRDS(INDEX)
#> }
#> else {
#> dataDir <- file.path(path, "data")
#> entries <- tools::list_files_with_type(dataDir,
#> "data")
#> if (length(entries)) {
#> entries <- unique(tools::file_path_sans_ext(basename(entries)))
#> entries <- cbind(entries, "")
#> }
#> }
#> if (NROW(entries)) {
#> if (is.matrix(entries) && ncol(entries) == 2L)
#> db <- rbind(db, cbind(packageName, dirname(path),
#> entries))
#> else warning(gettextf("data index for package %s is invalid and will be ignored",
#> sQuote(packageName)), domain = NA, call. = FALSE)
#> }
#> }
#> colnames(db) <- c("Package", "LibPath", "Item", "Title")
#> footer <- if (missing(package))
#> paste0("Use ", sQuote(paste("data(package =", ".packages(all.available = TRUE))")),
#> "\n", "to list the data sets in all *available* packages.")
#> else NULL
#> y <- list(title = "Data sets", header = NULL, results = db,
#> footer = footer)
#> class(y) <- "packageIQR"
#> return(y)
#> }
#> paths <- file.path(paths, "data")
#> for (name in names) {
#> found <- FALSE
#> for (p in paths) {
#> tmp_env <- if (overwrite)
#> envir
#> else new.env()
#> if (file_test("-f", file.path(p, "Rdata.rds"))) {
#> rds <- readRDS(file.path(p, "Rdata.rds"))
#> if (name %in% names(rds)) {
#> found <- TRUE
#> if (verbose)
#> message(sprintf("name=%s:\t found in Rdata.rds",
#> name), domain = NA)
#> thispkg <- sub(".*/([^/]*)/data$", "\\1", p)
#> thispkg <- sub("_.*$", "", thispkg)
#> thispkg <- paste0("package:", thispkg)
#> objs <- rds[[name]]
#> lazyLoad(file.path(p, "Rdata"), envir = tmp_env,
#> filter = function(x) x %in% objs)
#> break
#> }
#> else if (verbose)
#> message(sprintf("name=%s:\t NOT found in names() of Rdata.rds, i.e.,\n\t%s\n",
#> name, paste(names(rds), collapse = ",")),
#> domain = NA)
#> }
#> if (file_test("-f", file.path(p, "Rdata.zip"))) {
#> warning("zipped data found for package ", sQuote(basename(dirname(p))),
#> ".\nThat is defunct, so please re-install the package.",
#> domain = NA)
#> if (file_test("-f", fp <- file.path(p, "filelist")))
#> files <- file.path(p, scan(fp, what = "", quiet = TRUE))
#> else {
#> warning(gettextf("file 'filelist' is missing for directory %s",
#> sQuote(p)), domain = NA)
#> next
#> }
#> }
#> else {
#> files <- list.files(p, full.names = TRUE)
#> }
#> files <- files[grep(name, files, fixed = TRUE)]
#> if (length(files) > 1L) {
#> o <- match(fileExt(files), dataExts, nomatch = 100L)
#> paths0 <- dirname(files)
#> paths0 <- factor(paths0, levels = unique(paths0))
#> files <- files[order(paths0, o)]
#> }
#> if (length(files)) {
#> for (file in files) {
#> if (verbose)
#> message("name=", name, ":\t file= ...", .Platform$file.sep,
#> basename(file), "::\t", appendLF = FALSE,
#> domain = NA)
#> ext <- fileExt(file)
#> if (basename(file) != paste0(name, ".", ext))
#> found <- FALSE
#> else {
#> found <- TRUE
#> zfile <- file
#> zipname <- file.path(dirname(file), "Rdata.zip")
#> if (file.exists(zipname)) {
#> Rdatadir <- tempfile("Rdata")
#> dir.create(Rdatadir, showWarnings = FALSE)
#> topic <- basename(file)
#> rc <- .External(C_unzip, zipname, topic,
#> Rdatadir, FALSE, TRUE, FALSE, FALSE)
#> if (rc == 0L)
#> zfile <- file.path(Rdatadir, topic)
#> }
#> if (zfile != file)
#> on.exit(unlink(zfile))
#> switch(ext, R = , r = {
#> library("utils")
#> sys.source(zfile, chdir = TRUE, envir = tmp_env)
#> }, RData = , rdata = , rda = load(zfile,
#> envir = tmp_env), TXT = , txt = , tab = ,
#> tab.gz = , tab.bz2 = , tab.xz = , txt.gz = ,
#> txt.bz2 = , txt.xz = assign(name, my_read_table(zfile,
#> header = TRUE, as.is = FALSE), envir = tmp_env),
#> CSV = , csv = , csv.gz = , csv.bz2 = ,
#> csv.xz = assign(name, my_read_table(zfile,
#> header = TRUE, sep = ";", as.is = FALSE),
#> envir = tmp_env), found <- FALSE)
#> }
#> if (found)
#> break
#> }
#> if (verbose)
#> message(if (!found)
#> "*NOT* ", "found", domain = NA)
#> }
#> if (found)
#> break
#> }
#> if (!found) {
#> warning(gettextf("data set %s not found", sQuote(name)),
#> domain = NA)
#> }
#> else if (!overwrite) {
#> for (o in ls(envir = tmp_env, all.names = TRUE)) {
#> if (exists(o, envir = envir, inherits = FALSE))
#> warning(gettextf("an object named %s already exists and will not be overwritten",
#> sQuote(o)))
#> else assign(o, get(o, envir = tmp_env, inherits = FALSE),
#> envir = envir)
#> }
#> rm(tmp_env)
#> }
#> }
#> invisible(names)
#> }
#> <bytecode: 0x13f311aa8>
#> <environment: namespace:utils>
data1 <- data()
str(data1)
#> List of 4
#> $ title : chr "Data sets"
#> $ header : NULL
#> $ results: chr [1:104, 1:4] "datasets" "datasets" "datasets" "datasets" ...
#> ..- attr(*, "dimnames")=List of 2
#> .. ..$ : NULL
#> .. ..$ : chr [1:4] "Package" "LibPath" "Item" "Title"
#> $ footer : chr "Use 'data(package = .packages(all.available = TRUE))'\nto list the data sets in all *available* packages."
#> - attr(*, "class")= chr "packageIQR"
Created on 2023-06-26 with reprex v2.0.2
So on line 21 rename data to something like d that won't conflict. The rest is guesswork without a a reprex (see the FAQ). Screenshots are no substitute.
The function reactive takes an expression as its argument.
Reactive expressions let you control which parts of your app update when, which prevents unnecessary computation that can slow down your app. Shiny Tutorial
It's not clear that's happening here. After the logical test of req a df object is created, but doesn't appear to be returned as part of output. You probably want something along these lines
library(shiny)
library(readxl)
library(readr)
ui <- fluidPage(
titlePanel("File Reader"),
sidebarLayout(
sidebarPanel(
fileInput("file", "Choose a CSV or XLSX File")
),
mainPanel(
tableOutput("table")
)
)
)
server <- function(input, output) {
d <- reactive({
inFile <- input$file
if (is.null(inFile)) {
return(NULL)
}
ext <- tools::file_ext(inFile$datapath)
if (ext == "csv") {
read_csv(inFile$datapath)
} else if (ext == "xlsx") {
read_excel(inFile$datapath)
} else {
stop("Invalid file format. Please upload a CSV or XLSX file.")
}
})
output$table <- renderTable({
d()
})
}
shinyApp(ui = ui, server = server)
You are asking to do something which is simply against the shiny framework.
reactive data that you access via () within a reactive context is the heart and soul of a working shiny app. I recommend you adjust to that, rather than fight against it.
You're right, I couldn't agree with you more, I'll just find another way
Its not clear what problem if any you wish to solve ? I'm happy to try and give you advice if there is something you wish to accomplish and are blocked...
Thank you very much, I have solved it, I just simply use "data()" and not try to remove "()" to use it, thanks.
This topic was automatically closed 7 days after the last reply. New replies are no longer allowed.
If you have a query related to it or one of the replies, start a new topic and refer back with a link.