Yay!
Have you ever used summarise()?
It's a dplyr function in some ways similar to mutate, but with very important differences.
While mutate() will add a column/variable and keep all the observations, summarise() will take grouped data, return the columns/variables you create and collapse down to only the rows which are unique.
This is probably best seen with an example. You can copy and paste this and run it yourself so you can see what the resuling data looks like.
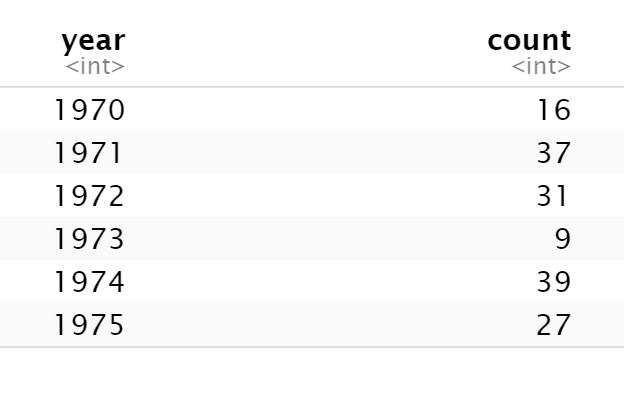
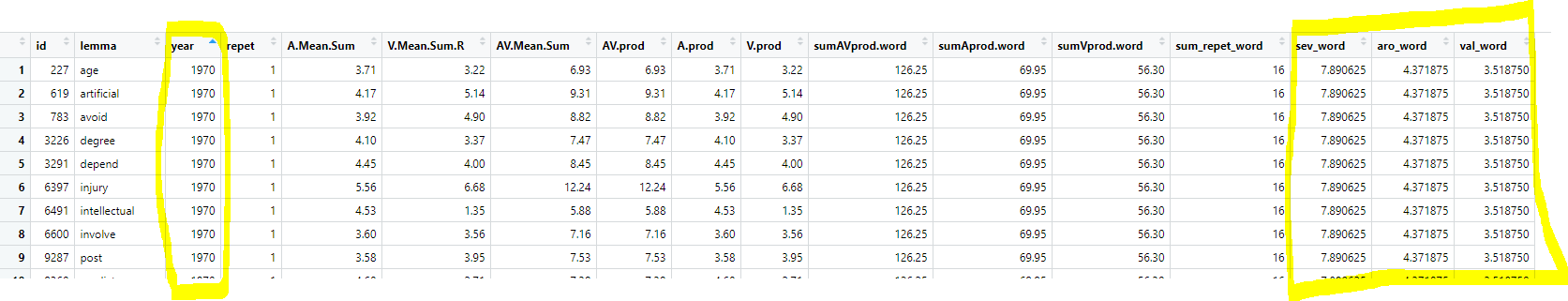
Here I create a datframe of 4,000 phone calls with 12 different customers.
calls <- tibble(callNo = 1:4000,
callDuration = as.integer(runif(4000, min = 60, max = 1200)),
customerNo = as.integer(runif(4000, min = 1, max =13)),
randomVar = runif(4000, min = 0, max = 1))
Now if I want some summary statistics, I can create them via mutate:
callSummary <- calls %>%
group_by(customerNo) %>%
mutate(callsPerCustomer = n(),
meanCallDur = sum(callDuration) / callsPerCustomer) %>%
ungroup()
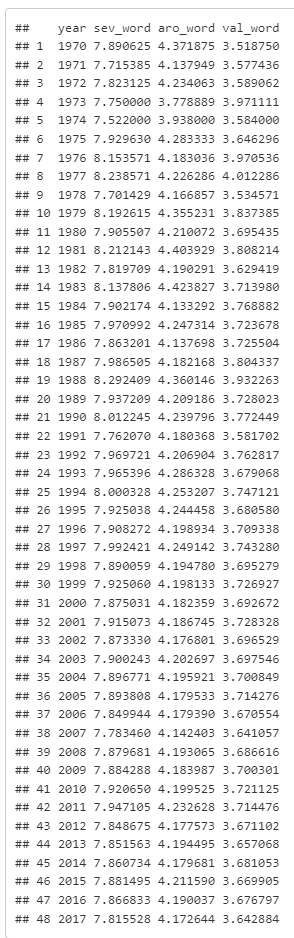
This gives the data I want, but keeps a lot that I no longer need.I still have 4,000 and some now useless columns. (So in reality, I'd add some code to select() the columns I want, then unique() to keep only the rows I need (eliminate now duplicate rows).
But check out summarise() (or summarize()):
callSummary <- calls %>%
group_by(customerNo) %>%
summarise(callsPerCustomer = n(),
meanCallDur = sum(callDuration) / callsPerCustomer) %>%
ungroup()
Same amount of code; I've only swapped summarise() in for mutate(), but this returns just 12 rows and 3 columns with just the data I was after.
I hope that helps. I have a project to finish this morning, so I just quickly read through your message, but it seems like what you might need. if you end up needing more help, please just ask.
Luke