I just upgraded to RStudio 2023.12.1 Build 402 and now my code throws this error. The code ran fine before the upgrade.
Markdown Notebooks
Tidyverse
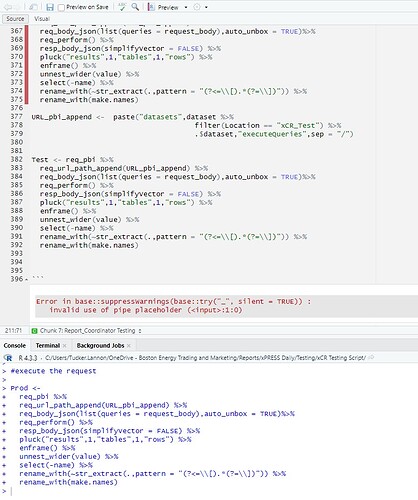
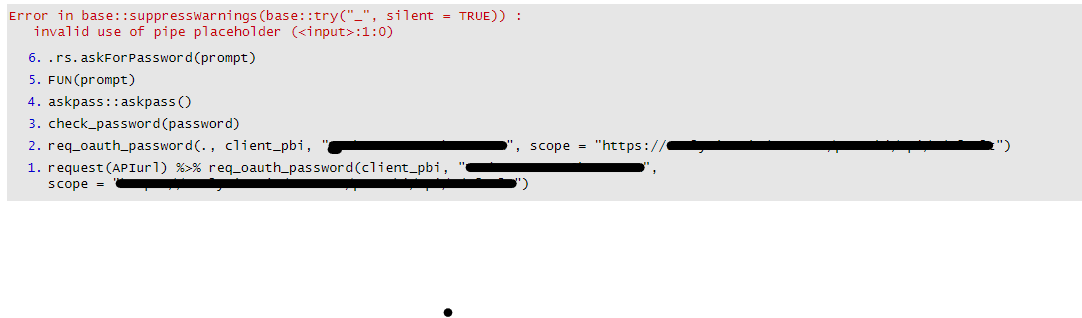
Error in base::suppressWarnings(base::try("_", silent = TRUE)) :
invalid use of pipe placeholder (:1:0)
Hi Tucker,
Could you share the code that caused the error, and send a screenshot of the output when you execute the code?
Hi The code above generates the error in in Rstudio markdown as you can see in the screen shot. However it does not generate the error in the console when you paste it in. The code uses httr2.
it seems like the errors are happening in the markdown and not in R.
Here is a function that generates the error inconsistently.
ClientReportingQuery <- function(query,location) {
request_body <- data.frame(query = c(query))
URL_pbi_append <- paste("datasets",dataset %>%
filter(Location == location) %>%
.$dataset,"executeQueries",sep = "/")results <- req_pbi %>%
req_url_path_append(URL_pbi_append) %>%
req_body_json(list(queries = request_body),auto_unbox = TRUE)%>%
req_perform() %>%
resp_body_json(simplifyVector = FALSE) %>%
pluck("results",1,"tables",1,"rows") %>%
enframe() %>%
unnest_wider(value) %>%
select(-name) %>%
rename_with(~str_extract(.,pattern = "(?<=\[).*(?=\])")) %>%
rename_with(make.names)results
}
This is the only place I can spot where something might be going on: The error is associated with the use of the base pipe (|>), not the magrittr pipe (%>%), so I'm wondering whether the R markdown code block is getting translated from magrittr to base, so to speak. (Piping to the $ operator would not work with the base pipe.)
What happens if you rewrite the offending code with the base pipe instead of the magrittr pipe?
- The error is also happening with the request component of the httr2 Auth process. This one is cool because its green for a few seconds while the password box pops up and then turns red and gives me the error below before i have entered my password and the code has finished running.
- Could you help me with the syntax on your earlier suggestion. I don't have a ton of base r experence.
Cheers
I just realized that this one is easy to rewrite, but from your later posts, it's not clear this would resolve the issue.
Could you make the following edit and see if it has any effect?
Replace this:
URL_pbi_append <- paste("datasets",dataset %>%
filter(Location == location) %>%
.$dataset,"executeQueries",sep = "/")
by this:
URL_pbi_append <-
paste(
"datasets",
dataset %>%
filter(Location == location) %>%
pull(dataset),
"executeQueries",
sep = "/"
)
I made the change and experienced the same results. it feels like there is something amiss with the way the markdown chunks work.
Just another thought: Did you update your R version, too? Or just your RStudio version?
I updated both Rstudio and R
Sorry — it's a mystery to me, but maybe other folks will have more insight. May be worth opening an issue in RStudio's GitHub site to get the developers' eyes on it.
Thanks for looking at it with me.
I can confirm that I'm getting the same issue in some knitr chunks. It is clearly something strange that is going on with knitr/rmarkdown/rstudio because the code runs fine on the console and it actually runs correctly in the chunk (i.e. the code outputs the result and is correct).
I am using the native pipe placeholder, but in a previous chunk. Furthermore, this issue appears only if I run all chunks above it and only the first time I do it. If I run each chunk by itself, it works well and if I click run all previous chunk twice, the second time it runs correctly.
I'm seeing this a lot as well.
My setup chunk often won't run, it errors out randomly even though the same code has worked in the past without issue.
The second chunk sources a simple script.
That script contains multiple library() to load the necessary packages for my analysis.
This error is repeated multiple times in the chunk output
Error in base::suppressWarnings(base::try("_", silent = TRUE)) :
invalid use of pipe placeholder (<input>:1:0)
The traceback shows the following
Error in base::suppressWarnings(base::try("_", silent = TRUE)) :
invalid use of pipe placeholder (<input>:1:0)
7.
apply(!is.na(db), 1L, all)
6.
find.package(package, lib.loc, quiet = TRUE, verbose = verbose)
5.
library(dplyr) at 02-packages.R#5
4.
eval(ei, envir)
3.
eval(ei, envir)
2.
withVisible(eval(ei, envir))
1.
source("./rmd-processing/02-packages.R")
And so on- this base pipe error keeps cropping up, even for code where there are no pipes being used.
Code that should run freely within the chunk seems to trip over and error out for no obvious reason.
R version 4.3.1 (2023-06-16 ucrt)
R Studio 2023.12.1 Build 402
Windows
This topic was automatically closed 21 days after the last reply. New replies are no longer allowed.
If you have a query related to it or one of the replies, start a new topic and refer back with a link.