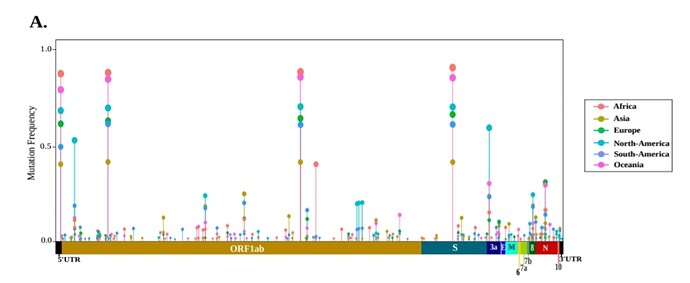
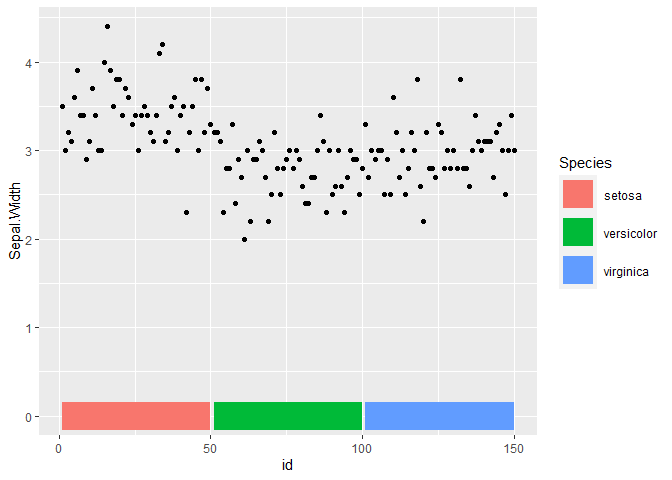
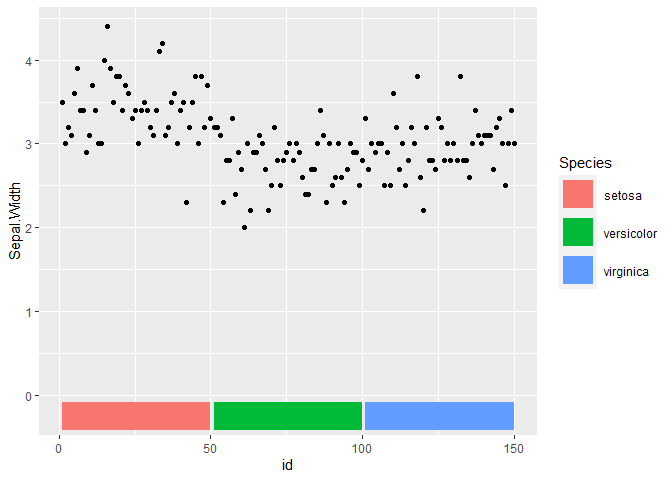
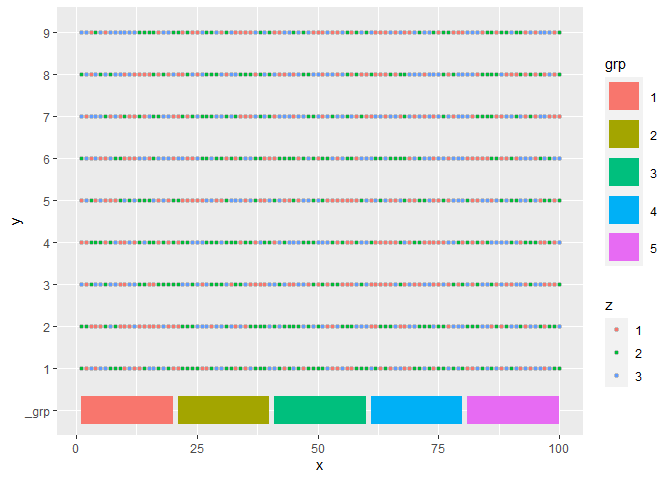
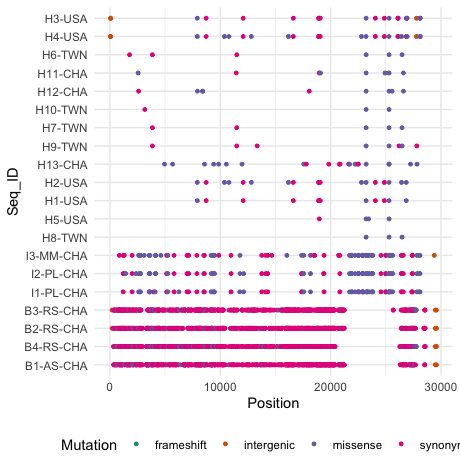
I'm analyzing SARS variants, which I manipulated the data with tidyverse and acquired the following dataset
> glimpse(data)
Rows: 2,861
Columns: 5
$ Position <int> 67, 67, 70, 70, 72, 72, 1782, 2557, 2601, 3165, 3852, 3852, 3852, 4952, 5681, 7919, 7919, 7919, 7919, 7930…
$ ORF <chr> "5UTR", "5UTR", "5UTR", "5UTR", "5UTR", "5UTR", "ORF1a", "ORF1a", "ORF1a", "ORF1a", "ORF1a", "ORF1a", "ORF…
$ Mutation <chr> "intergenic", "intergenic", "intergenic", "intergenic", "intergenic", "intergenic", "synonymous", "missens…
$ Seq_ID <chr> "H3-USA", "H4-USA", "H3-USA", "H4-USA", "H3-USA", "H4-USA", "H6-TWN", "H11-CHA", "H12-CHA", "H10-TWN", "H6…
$ Host <chr> "Human", "Human", "Human", "Human", "Human", "Human", "Human", "Human", "Human", "Human", "Human", "Human"...
From this data I made the follow figure using the ggplot2 and the script employed is the one shown below:
data$Seq_ID <- factor(data$Seq_ID, levels = rev(unique(data$Seq_ID)))
ggplot(data) + geom_point(aes(Position, Seq_ID, color = Mutation), size = 1) + theme_minimal() + theme(legend.position = "bottom") + scale_color_brewer(palette = "Dark2")
Rplot01.pdf (122.3 KB)
Visually it looks like what I want. However, several aspects are still to be annotated according to the figure I show below
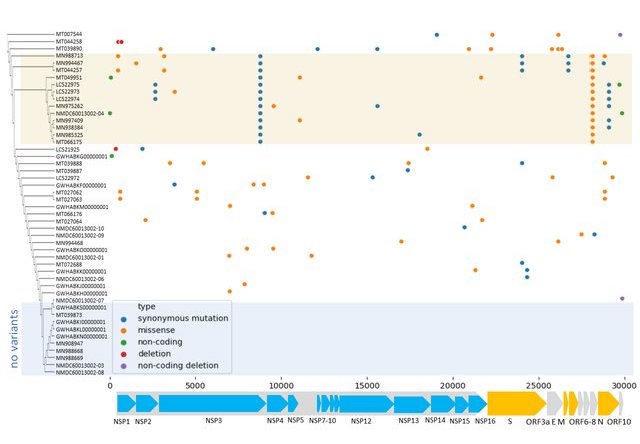
These are the tasks I need help:
- To each type of host assign a different color as background (bat, intermediate, human) that is seen in the Y-axis.
- Build the bar chart near the X-axis, where it shows the different ORFs.
I appreciate very much any help you can contribute.