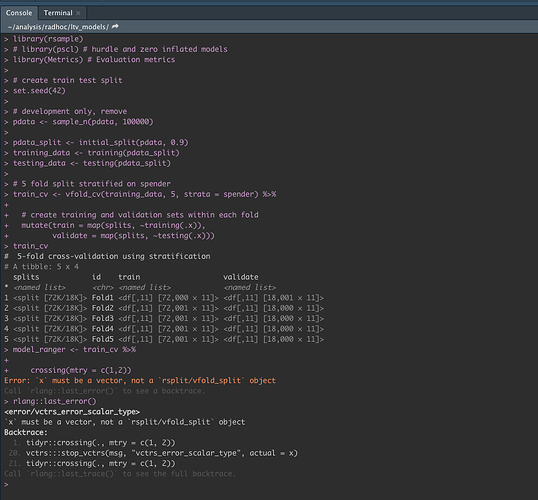
Am encountering the above error message when attempting to apply crossing() to what I think a nested data frame, but I'm not sure:
model_ranger <- train_cv %>%
crossing(mtry = c(1,2)) # %>%
Sometimes results in:
Error:
xmust be a vector, not arsplit/vfold_splitobject
This error happens only sometimes, if I just keep re running the code block it sometimes works. (Discovered by a combination of accident and desperation).
The object in question:
> class(train_cv)
[1] "vfold_cv" "rset" "tbl_df" "tbl" "data.frame"
> train_cv
# 5-fold cross-validation using stratification
# A tibble: 5 x 4
splits id train validate
* <named list> <chr> <named list> <named list>
1 <split [72K/18K]> Fold1 <df[,11] [72,000 × 11]> <df[,11] [18,001 × 11]>
2 <split [72K/18K]> Fold2 <df[,11] [72,001 × 11]> <df[,11] [18,000 × 11]>
3 <split [72K/18K]> Fold3 <df[,11] [72,001 × 11]> <df[,11] [18,000 × 11]>
4 <split [72K/18K]> Fold4 <df[,11] [72,001 × 11]> <df[,11] [18,000 × 11]>
5 <split [72K/18K]> Fold5 <df[,11] [72,001 × 11]> <df[,11] [18,000 × 11]>
I arrived here with the following block of code, where pdata is my starting point regular df.
library(rsample)
# create train test split
set.seed(123)
pdata_split <- initial_split(pdata, 0.9)
training_data <- training(pdata_split)
testing_data <- testing(pdata_split)
# 5 fold split stratified on spender
train_cv <- vfold_cv(training_data, 5, strata = spender) %>%
# create training and validation sets within each fold
mutate(train = map(splits, ~training(.x)),
validate = map(splits, ~testing(.x)))
Now that I've created the splits, I have separate code blocks that fit a ranger random forrest and an xgb on the same folds. For ranger I start with:
model_ranger <- train_cv %>%
crossing(mtry = c(1,2)) # %>%
Error:
xmust be a vector, not arsplit/vfold_splitobject
I tried to recreate this using diamonds built in dataset, but it worked. It's just with my actual data this happens. Intermittently.
Any ideas on how to solve?