My data is a CSV file from Microsoft Excel. This file contains several numeric variables (e.g. salary, weight) that are written in the following format: 123,456 (one hundred and twenty three thousand four hundred and fifty six).
When I import this file ("old_file") into R using the following command : my_file = read.csv("old_file.csv") , all the numeric variables are automatically converted to factor variable types.
To solve this problem, I tried to manually convert these variables into numeric variable types after importing the file into R:
my_file$var_1 = as.numeric(my_file$var_1)
However, doing this seems to "distort" the data. For example, values of "var_1" that are naturally occurring 0's seem to disappear. I also plotted some histograms of this data and the distribution of these numeric converted variables do not match the expected distributions.
I was able to solve this problem by opening the Excel Spreadsheet and clicking the "format button" and removing the "commas" from the numeric variables, and adding ".00" to the end of each number. For example, 123,456 now becomes 123456.00 .
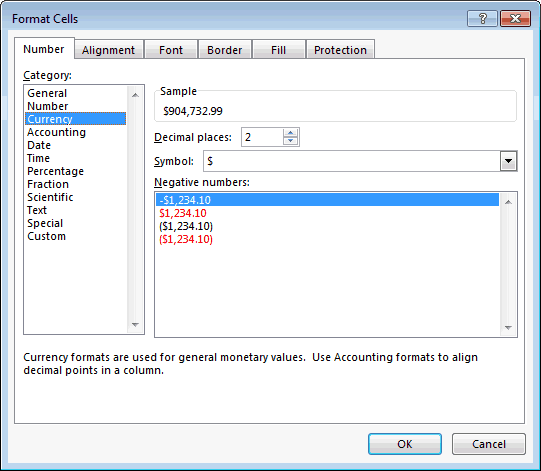
Once I made this change in the original Excel Spreadsheet, the problem was then resolved.
My Question: Is there a way to solve this problem in R, instead of manually doing this in the original Excel Spreadsheet? I just happened to catch this formatting error - in the future, is there a way to automatically prevent this from happening in R? E.g. Some way to ensure that variables that "appear to be numeric in the Excel File" get imported as numeric types in R?