Hello,
I have 6 SNPs of 3 gene regions that are in linkage desequilibrium (highly correlated) and i would like to identify haplotypes to further used them in association study instead of the individual SNP. I found that the function haplo.em from the haplo.stats packages allows to identify the haplotypes and their probability (see haplostats.pdf (r-project.org) section 3 - page 8). I have some questions regarding the data that we should load in R to use this function and the interpretation of the results :
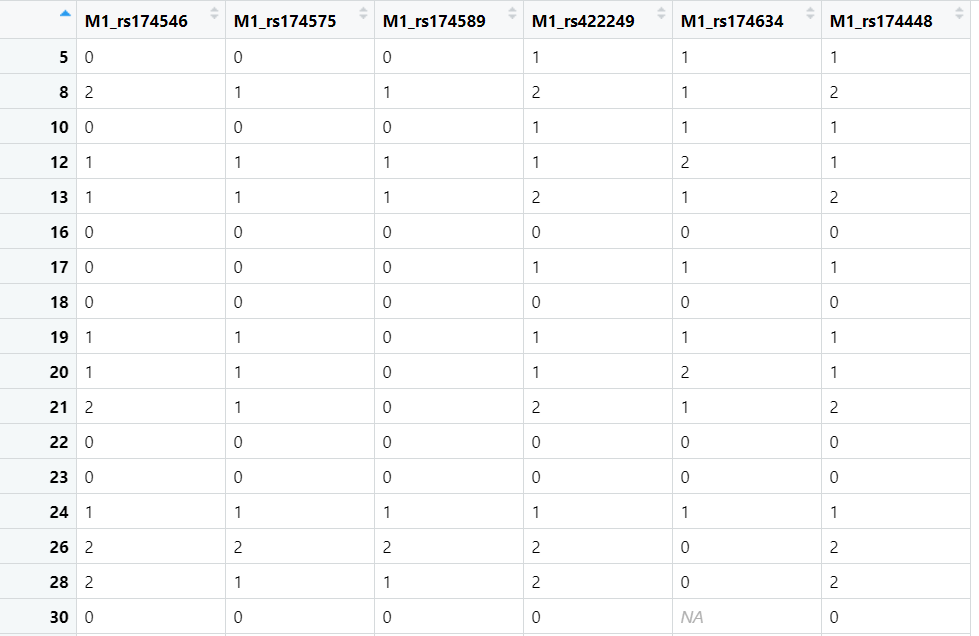
- What format of the data should we load ? Could it be a matrix corresponding in rows to the individuals and a column for each SNP (see the attached file data.png attached) with 0 = major homozygote, 1= heterozygote, 2=minor homozygote ? or should it be as Aa, AA or aa directy ? or something else ?
- I dont understand the table of results (section 3.2 "Example Usage") in the Rdocumentation ? how are we suppose to know the combination of alleles corresponding to each haplotype ? Except the last column that is the frequency, i dont get what the other columns give us.
- Is there a way to obtain a table with the different haplotypes identified, their frequency and for each one, the corresponding combination of alleles for the SNPs ?
The genetic world is something new to me and dealing with it on R is even harder. I would appreciate any help.
Thank you in advance !
Have a nice day,
Aline