Dear community,
Quite a few months ago I wrote this code for importing a csv file that worked fine at the time :
library(tidyverse)
###################################################################################### p 1000 t1 #################
# read each row of text individually so we can parse out the information manually
election0 <-
read_delim(
"~/MASTER_1/stage/travail/R/donnees/2014/2014_t1+1000.txt",
"\n",
col_names = FALSE,locale = locale(encoding = "ISO-8859-1")) %>%
setNames("line_text") %>%
mutate(
# split by delimiter
split_text = strsplit(line_text, ";"),
# assume the first 17 elements are common
split_df = map(split_text, ~.[1:17]),
# and everything past this is repeating 11
split_names = map(split_text, ~.[-c(1:17)]),
columns = map_dbl(split_text, length),
# the number of repeating 11 name data elements
n_names = (columns - 17)/11)
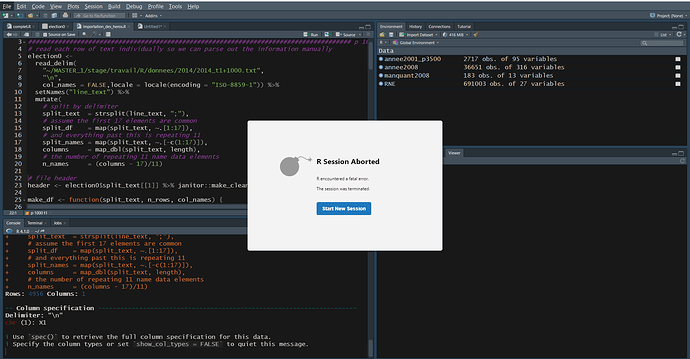
But when I run this same code on the same data as before my IDE crash (I think) I get a dialog box saying
R session aborded, R encountered a fatal error. The session was terminated :
Here is my session info :
> sessionInfo()
R version 4.1.0 (2021-05-18)
Platform: x86_64-w64-mingw32/x64 (64-bit)
Running under: Windows 10 x64 (build 19043)
Matrix products: default
locale:
[1] LC_COLLATE=French_France.1252 LC_CTYPE=French_France.1252
[3] LC_MONETARY=French_France.1252 LC_NUMERIC=C
[5] LC_TIME=French_France.1252
attached base packages:
[1] stats graphics grDevices utils datasets methods base
other attached packages:
[1] forcats_0.5.1 stringr_1.4.0 dplyr_1.0.7 purrr_0.3.4 readr_2.0.0
[6] tidyr_1.1.3 tibble_3.1.3 ggplot2_3.3.5 tidyverse_1.3.1
loaded via a namespace (and not attached):
[1] Rcpp_1.0.7 cellranger_1.1.0 pillar_1.6.2 compiler_4.1.0 dbplyr_2.1.1
[6] tools_4.1.0 lubridate_1.7.10 jsonlite_1.7.2 lifecycle_1.0.0 gtable_0.3.0
[11] pkgconfig_2.0.3 rlang_0.4.11 reprex_2.0.1 cli_3.0.1 rstudioapi_0.13
[16] DBI_1.1.1 haven_2.4.3 xml2_1.3.2 withr_2.4.2 httr_1.4.2
[21] fs_1.5.0 generics_0.1.0 vctrs_0.3.8 hms_1.1.0 grid_4.1.0
[26] tidyselect_1.1.1 glue_1.4.2 R6_2.5.0 fansi_0.5.0 readxl_1.3.1
[31] tzdb_0.1.2 modelr_0.1.8 magrittr_2.0.1 backports_1.2.1 scales_1.1.1
[36] ellipsis_0.3.2 rvest_1.0.1 assertthat_0.2.1 colorspace_2.0-2 utf8_1.2.2
[41] stringi_1.7.3 munsell_0.5.0 broom_0.7.9 crayon_1.4.1
And an extract of the logfile :
09 Aug 2021 07:03:02 [rsession-zugat] ERROR system error 10053 (Une connexion établie a été abandonnée par un logiciel de votre ordinateur hôte) [request-uri: /events/get_events]; OCCURRED AT void __cdecl rstudio::session::HttpConnectionImpl<class rstudio_boost::asio::ip::tcp>::sendResponse(const class rstudio::core::http::Response &) src/cpp/session/http/SessionWin32HttpConnectionListener.cpp:113; LOGGED FROM: void __cdecl rstudio::session::HttpConnectionImpl<class rstudio_boost::asio::ip::tcp>::sendResponse(const class rstudio::core::http::Response &) src/cpp/session/http/SessionWin32HttpConnectionListener.cpp:118
09 Aug 2021 07:18:18 [rsession-zugat] ERROR system error 5 (Accès refusé); OCCURRED AT auto __cdecl rstudio::core::system::ChildProcess::terminate::<lambda_b34d56978c1a268cda78ea8a24bc0d35>::operator ()(void) const src/cpp/core/system/Win32ChildProcess.cpp:287; LOGGED FROM: void __cdecl rstudio::core::system::ProcessSupervisor::terminateAll(void) src/cpp/core/system/Process.cpp:363
It's the first time I see the second error since I'm trying to execute the code so i don't think that one is causing trouble but I don't know.
Your help is much appreciated, Best Regards