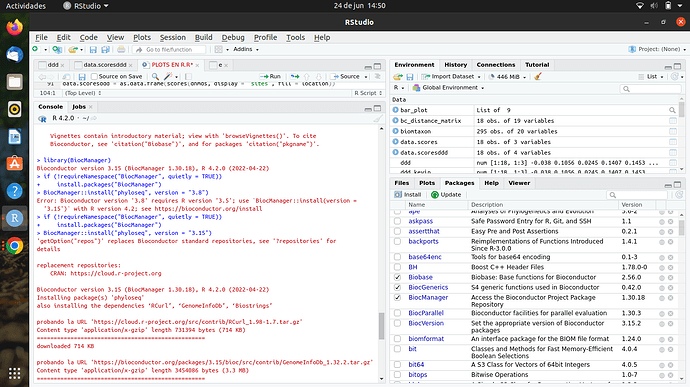
hi, im new into R, im doing my first steps and i want to make an analysis of microbiome with phyloseq package, im working on ubuntu 20.04 LTS, with R version 4.2.0 (2022-04-22) -- "Vigorous Calisthenics"
but when Im trying to install the package, Rstudio shows me the following:
if (!requireNamespace("BiocManager", quietly = TRUE))
-
install.packages("BiocManager")
BiocManager::install("phyloseq", version = "3.15")
'getOption("repos")' replaces Bioconductor standard repositories, see '?repositories' for
details
replacement repositories:
CRAN: https://cloud.r-project.org
Bioconductor version 3.15 (BiocManager 1.30.18), R 4.2.0 (2022-04-22)
Installing package(s) 'phyloseq'
also installing the dependencies ‘RCurl’, ‘GenomeInfoDb’, ‘Biostrings’
- installing source package ‘RCurl’ ...
** package ‘RCurl’ successfully unpacked and MD5 sums checked
** using staged installation
checking for curl-config... no
Cannot find curl-config
ERROR: configuration failed for package ‘RCurl’ - removing ‘/home/kevin/R/x86_64-pc-linux-gnu-library/4.2/RCurl’
ERROR: dependency ‘RCurl’ is not available for package ‘GenomeInfoDb’ - removing ‘/home/kevin/R/x86_64-pc-linux-gnu-library/4.2/GenomeInfoDb’
ERROR: dependency ‘GenomeInfoDb’ is not available for package ‘Biostrings’ - removing ‘/home/kevin/R/x86_64-pc-linux-gnu-library/4.2/Biostrings’
ERROR: dependency ‘Biostrings’ is not available for package ‘phyloseq’ - removing ‘/home/kevin/R/x86_64-pc-linux-gnu-library/4.2/phyloseq’
The downloaded source packages are in
‘/tmp/RtmpiV8M3T/downloaded_packages’
Installation paths not writeable, unable to update packages
path: /usr/lib/R/library
packages:
nlme, spatial, survival
Warning messages:
1: In install.packages(...) :
installation of package ‘RCurl’ had non-zero exit status
2: In install.packages(...) :
installation of package ‘GenomeInfoDb’ had non-zero exit status
3: In install.packages(...) :
installation of package ‘Biostrings’ had non-zero exit status
4: In install.packages(...) :
installation of package ‘phyloseq’ had non-zero exit status
do yuo know how to solve this issue?
Thanks