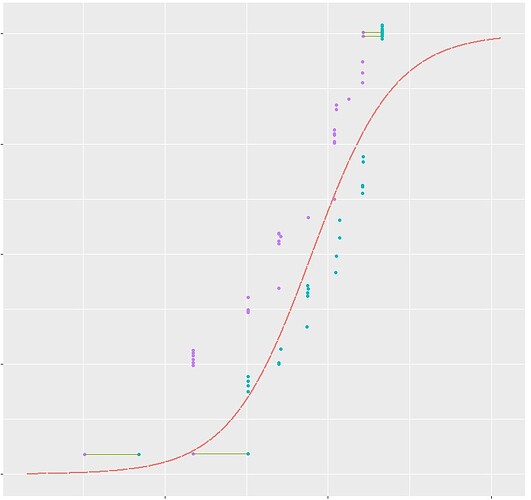
I want to link two points together using geom segment, could anyone please help me why I am not getting the lines?
Code has been attached below and attaching figure as well. Also, the points are not parallel as well.
structure(list(X = c(0.333, 133.333, 9999, 9999, 423.333, 123.333,
33.333, 1623.333, 9999, 33.333, 33.333, 1398.333, 33.318, 1623.048,
9999, 9999, 9999, 4323.333, 9999, 33.333, 408.108, 9999, 9999,
4323.333, 123.333, 4323.333, 1433.333, 433.333, 33.333, 4433.333,
423.333, 9999, 4433.333, 423.333), SO_obj_LOAEL = c(0.033, 33.333,
4433.333, 4433.333, 123.333, 33.333, 3.333, 1323.333, 4285.833,
3.333, 3.333, 1323.333, 3.318, 1323.048, 4323.333, 3.333, 4323.108,
1323.333, 33.333, 3.333, 123.108, 123.333, 3.333, 1323.333, 33.333,
1323.333, 433.333, 133.333, 3.333, 1433.333, 123.333, 2433.333,
1433.333, 123.333), SO_obj_NOAEL = c(1L, 1L, NA, NA, 1L, 1L,
1L, 1L, NA, 1L, 1L, 1L, 1L, 1L, NA, NA, NA, 1L, NA, 1L, 1L, NA,
NA, 1L, 1L, 1L, 1L, 1L, 1L, 1L, 1L, NA, 1L, 1L), SO_OUTCOME_obj = c(3L,
3L, NA, NA, 3L, 3L, 3L, 3L, NA, 3L, 3L, 3L, 3L, 3L, NA, NA, NA,
3L, NA, 3L, 3L, NA, NA, 3L, 3L, 3L, 3L, 3L, 3L, 3L, 3L, NA, 3L,
3L), SO_CENSORING_obj = c(0.333, 133.333, 9999, 9999, 423.333,
123.333, 33.333, 1623.333, 9999, 33.333, 33.333, 1398.333, 33.318,
1623.048, 9999, 9999, 9999, 4323.333, 9999, 33.333, 408.108,
9999, 9999, 4323.333, 123.333, 4323.333, 1433.333, 433.333, 33.333,
4433.333, 423.333, 9999, 4433.333, 423.333), modified_SO_obj_LOAEL = c(0.033,
33.333, 4433.333, 4433.333, 123.333, 33.333, 3.333, 1323.333,
4285.833, 3.333, 3.333, 1323.333, 3.318, 1323.048, 4323.333,
3.333, 4323.108, 1323.333, 33.333, 3.333, 123.108, 123.333, 3.333,
1323.333, 33.333, 1323.333, 433.333, 133.333, 3.333, 1433.333,
123.333, 2433.333, 1433.333, 123.333), modified_SO_obj_NOAEL = structure(c(4L,
7L, 19L, 19L, 9L, 7L, 6L, 13L, 16L, 6L, 6L, 13L, 5L, 12L, 18L,
1L, 17L, 13L, 2L, 6L, 8L, 3L, 1L, 13L, 7L, 13L, 11L, 10L, 6L,
14L, 9L, 15L, 14L, 9L), .Label = c(" 3.333+ ", " 33.333+ ",
" 123.333+ ", "[ 0.033", "[ 3.318", "[ 3.333",
"[ 33.333", "[ 123.108", "[ 123.333", "[ 133.333", "[ 433.333",
"[1323.048", "[1323.333", "[1433.333", "2433.333+ ",
"4285.833+ ", "4323.108+ ", "4323.333+ ",
"4433.333+ "), class = "factor"), Survial_analysis_obj = structure(c(1L,
5L, 22L, 22L, 7L, 4L, 3L, 12L, 19L, 3L, 3L, 9L, 2L, 11L, 21L,
15L, 20L, 13L, 16L, 3L, 6L, 17L, 15L, 13L, 4L, 13L, 10L, 8L,
3L, 14L, 7L, 18L, 14L, 7L), .Label = c(" 0.333]", " 33.318]",
" 33.333]", " 123.333]", " 133.333]", " 408.108]", " 423.333]",
" 433.333]", " 1398.333]", " 1433.333]", " 1623.048]", " 1623.333]",
" 4323.333]", " 4433.333]", "0.26470588", "0.38235294", "0.52941176",
"0.85294118", "0.88235294", "0.91176471", "0.94117647", "1.00000000"
), class = "factor"), NOAELcdf = c(0.02941176, 0.38235294, 1,
1, 0.52941176, 0.38235294, 0.26470588, 0.76470588, 1, 0.26470588,
0.26470588, 0.76470588, 0.05882353, 0.61764706, 1, 1, 1, 0.76470588,
1, 0.26470588, 0.41176471, 1, 1, 0.76470588, 0.38235294, 0.76470588,
0.58823529, 0.55882353, 0.26470588, 0.82352941, 0.52941176, 1,
0.82352941, 0.52941176), LOAELcdf = c(0.02941176, 0.29411765,
-0.005115044, 0.002914135, 0.41176471, 0.26470588, 0.20588235,
0.55882353, 0.006431912, 0.20588235, 0.20588235, 0.47058824,
0.05882353, 0.52941176, -0.004635851, 0.01079366, -9.203032e-05,
0.64705882, 0.01967624, 0.20588235, 0.32352941, 0.01738821, -0.0115143,
0.64705882, 0.26470588, 0.64705882, 0.5, 0.44117647, 0.20588235,
0.70588235, 0.41176471, -0.0007167954, 0.70588235, 0.41176471
), jit = c(0.01586789, -0.009379653, NA, NA, 0.01632831, -0.01193272,
0.01593559, 0.01778701, NA, 0.005164562, -0.01752855, -0.01176102,
-0.01293773, 0.007480914, NA, NA, NA, 0.00870474, NA, -0.004798593,
0.01109781, NA, NA, 0.006066951, -0.0149778, -0.009311173, -0.004555436,
-0.01946439, -0.004704482, 0.01478763, -0.00638604, NA, 0.003982633,
-0.0002583477)), .Names = c("X", "SO_obj_LOAEL", "SO_obj_NOAEL",
"SO_OUTCOME_obj", "SO_CENSORING_obj", "modified_SO_obj_LOAEL",
"modified_SO_obj_NOAEL", "Survial_analysis_obj", "NOAELcdf",
"LOAELcdf", "jit"), class = "data.frame", row.names = c("1",
"2", "3", "4", "5", "6", "7", "8", "9", "10", "11", "12", "13",
"14", "15", "16", "17", "18", "19", "20", "21", "22", "23", "24",
"25", "26", "27", "28", "29", "30", "31", "32", "33", "34"))
food<-read.csv("food.csv",header = TRUE,na.strings=0)
set.seed(0)
mydata$jit <- runif(nrow(mydata), -0.02, 0.02)
ggplot() +
geom_point(data = mydata, aes(x = as.numeric(lower), y = NOAELcdf + mydata$jit, colour = "lower")) +
geom_point(data = mydata, aes(x = as.numeric(higher), y = LOAELcdf + mydata$jit, colour = "higher")) +
geom_segment(aes(x = as.numeric(lower), y = ifelse(NOAELcdf!= LOAELcdf, NA, NOAELcdf)+ mydata$jit, xend = as.numeric(higher), yend = LOAELcdf + mydata$jit, colour = "segment"), data = mydata)+
######### Add in loglogistic curves
geom_line(data = predmodel_log_L_obj.curves,
aes(x = LogLogisticFit, y = probability,
colour = "LogLogisticFit"), size=1, linetype = "F1") +
########adding log scale to the x-axis
scale_x_log10() +
scale_y_continuous()