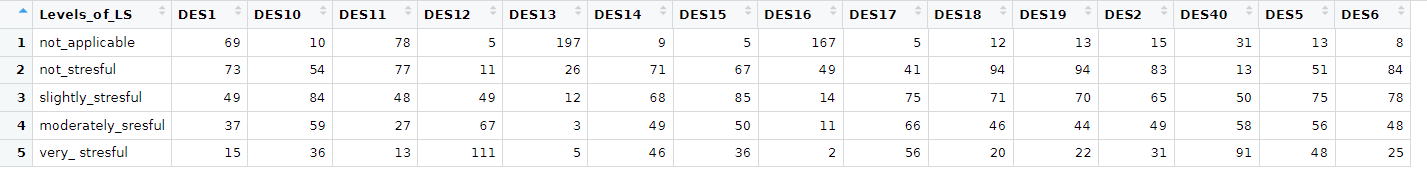
For each of the questions, the totals can be expanded. And the answers are mutually exclusive. So each row represents one subject. I don't think, though, that the results can be lined up.
d <- structure(list(
Levels_of_LS = c(
"not_applicable", "not_stresful",
"slightly_stresful", "moderately_sresful", "very_ stresful"
),
DES1 = c(69, 73, 49, 37, 15), DES10 = c(10, 54, 84, 59, 36), DES11 = c(78, 77, 48, 27, 13), DES12 = c(
5, 11, 49, 67,
111
), DES13 = c(197, 26, 12, 3, 5), DES14 = c(
9, 71, 68,
49, 46
), DES15 = c(5, 67, 85, 50, 36), DES16 = c(
167, 49,
14, 11, 2
), DES17 = c(5, 41, 75, 66, 56), DES18 = c(
12, 94,
71, 46, 20
), DES19 = c(13, 94, 70, 44, 22), DES2 = c(
15,
83, 65, 49, 31
), DES40 = c(31, 13, 50, 58, 91), DES5 = c(
13,
51, 75, 56, 48
), DES6 = c(8, 84, 78, 48, 25)
), class = c(
"tbl_df",
"tbl", "data.frame"
), row.names = c(NA, -5L))
dt <- as.data.frame(matrix(as.integer(t(d[,-1])), nrow = 15, ncol = 5))
dt$des <- colnames(d)[-1]
head(dt)
#> V1 V2 V3 V4 V5 des
#> 1 69 73 49 37 15 DES1
#> 2 10 54 84 59 36 DES10
#> 3 78 77 48 27 13 DES11
#> 4 5 11 49 67 111 DES12
#> 5 197 26 12 3 5 DES13
#> 6 9 71 68 49 46 DES14
example <- unlist(dt[,1])
head(example)
#> [1] 69 10 78 5 197 9
enlarge <- function(x) rep(1,x)
sapply(example,enlarge)
#> [[1]]
#> [1] 1 1 1 1 1 1 1 1 1 1 1 1 1 1 1 1 1 1 1 1 1 1 1 1 1 1 1 1 1 1 1 1 1 1 1 1 1 1
#> [39] 1 1 1 1 1 1 1 1 1 1 1 1 1 1 1 1 1 1 1 1 1 1 1 1 1 1 1 1 1 1 1
#>
#> [[2]]
#> [1] 1 1 1 1 1 1 1 1 1 1
#>
#> [[3]]
#> [1] 1 1 1 1 1 1 1 1 1 1 1 1 1 1 1 1 1 1 1 1 1 1 1 1 1 1 1 1 1 1 1 1 1 1 1 1 1 1
#> [39] 1 1 1 1 1 1 1 1 1 1 1 1 1 1 1 1 1 1 1 1 1 1 1 1 1 1 1 1 1 1 1 1 1 1 1 1 1 1
#> [77] 1 1
#>
#> [[4]]
#> [1] 1 1 1 1 1
#>
#> [[5]]
#> [1] 1 1 1 1 1 1 1 1 1 1 1 1 1 1 1 1 1 1 1 1 1 1 1 1 1 1 1 1 1 1 1 1 1 1 1 1 1
#> [38] 1 1 1 1 1 1 1 1 1 1 1 1 1 1 1 1 1 1 1 1 1 1 1 1 1 1 1 1 1 1 1 1 1 1 1 1 1
#> [75] 1 1 1 1 1 1 1 1 1 1 1 1 1 1 1 1 1 1 1 1 1 1 1 1 1 1 1 1 1 1 1 1 1 1 1 1 1
#> [112] 1 1 1 1 1 1 1 1 1 1 1 1 1 1 1 1 1 1 1 1 1 1 1 1 1 1 1 1 1 1 1 1 1 1 1 1 1
#> [149] 1 1 1 1 1 1 1 1 1 1 1 1 1 1 1 1 1 1 1 1 1 1 1 1 1 1 1 1 1 1 1 1 1 1 1 1 1
#> [186] 1 1 1 1 1 1 1 1 1 1 1 1
#>
#> [[6]]
#> [1] 1 1 1 1 1 1 1 1 1
#>
#> [[7]]
#> [1] 1 1 1 1 1
#>
#> [[8]]
#> [1] 1 1 1 1 1 1 1 1 1 1 1 1 1 1 1 1 1 1 1 1 1 1 1 1 1 1 1 1 1 1 1 1 1 1 1 1 1
#> [38] 1 1 1 1 1 1 1 1 1 1 1 1 1 1 1 1 1 1 1 1 1 1 1 1 1 1 1 1 1 1 1 1 1 1 1 1 1
#> [75] 1 1 1 1 1 1 1 1 1 1 1 1 1 1 1 1 1 1 1 1 1 1 1 1 1 1 1 1 1 1 1 1 1 1 1 1 1
#> [112] 1 1 1 1 1 1 1 1 1 1 1 1 1 1 1 1 1 1 1 1 1 1 1 1 1 1 1 1 1 1 1 1 1 1 1 1 1
#> [149] 1 1 1 1 1 1 1 1 1 1 1 1 1 1 1 1 1 1 1
#>
#> [[9]]
#> [1] 1 1 1 1 1
#>
#> [[10]]
#> [1] 1 1 1 1 1 1 1 1 1 1 1 1
#>
#> [[11]]
#> [1] 1 1 1 1 1 1 1 1 1 1 1 1 1
#>
#> [[12]]
#> [1] 1 1 1 1 1 1 1 1 1 1 1 1 1 1 1
#>
#> [[13]]
#> [1] 1 1 1 1 1 1 1 1 1 1 1 1 1 1 1 1 1 1 1 1 1 1 1 1 1 1 1 1 1 1 1
#>
#> [[14]]
#> [1] 1 1 1 1 1 1 1 1 1 1 1 1 1
#>
#> [[15]]
#> [1] 1 1 1 1 1 1 1 1