Hi, here is the code:
vec <- c(6, 3, 0, 9, 5)
for(i in 1:length(vec)) {
out <- vec[i] + 10
print(out)
}
for(i in 1:length(vec)) {
out[i] <- vec[i] + 10
print(out)
}
Desired output:
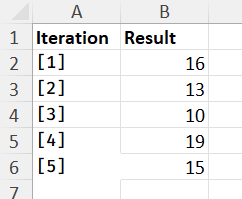
and:
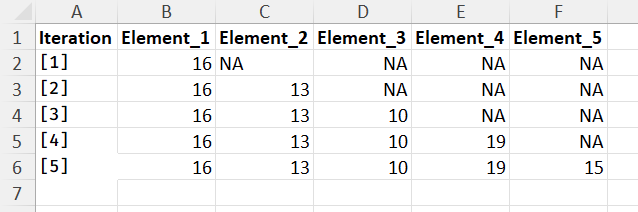
Any help much appreciated.
Hi, here is the code:
vec <- c(6, 3, 0, 9, 5)
for(i in 1:length(vec)) {
out <- vec[i] + 10
print(out)
}
for(i in 1:length(vec)) {
out[i] <- vec[i] + 10
print(out)
}
Desired output:

and:

Any help much appreciated.
Hey, what exactly do you need there? The "iteration" column is no where in your for-loop, so I guess it is not really necessary (please provide actual data instead of a picture if it would be necessary)?
Either way, your first result is just a vector, so instead of a for-loop you could just output vec + 10, which will add 10 to every element of your vector.
The second one can be achieved if you specify the exact position in a matrix:
vec <- c(6L, 3L, 0L, 9L, 5L)
out <- matrix(data = NA_integer_, nrow = length(vec), ncol = length(vec))
for (i in seq.default(1, length(vec))) {
out[i:length(vec), i] <- vec[[i]] + 10
}
> out
[,1] [,2] [,3] [,4] [,5]
[1,] 16 NA NA NA NA
[2,] 16 13 NA NA NA
[3,] 16 13 10 NA NA
[4,] 16 13 10 19 NA
[5,] 16 13 10 19 15
Kind regards
I have provided all data needed here. Thank you for your help.
For first part I have tried:
all_iter <- data.frame()
vec <- c(6, 3, 0, 9, 5)
for(i in 1:length(vec)) {
result <- vec[i] + 10
print(result)
all_iter <- all_iter %>% rbind(c(i, result))
}
I would like to better get to know how to write more advanced for loops and this is why I want to store each iteration step separately to see how does it work.
Additionally I want to understand when to use:
result <- vec[i] + 10, or result[1] <- vec[i] + 10 or result[[1]] <- vec[i] + 10
There is an excellent book from Hadley Wickham about R which covers the subsetting of R objects as well as other interesting topics. I think you will find what you need there.
I know this book and I have read it many times, but subsetting and assigning inside a for loop is a different fairy tale.
vec <- c(6, 3, 0, 9, 5)
(ten <- vec + 10)
#> [1] 16 13 10 19 15
m <- matrix(NA,nrow = 5,ncol = 5)
the_rows <- dim(m)[1]
for (i in 1:length(ten)) m[i:the_rows,i] <- ten[i]
m
#> [,1] [,2] [,3] [,4] [,5]
#> [1,] 16 NA NA NA NA
#> [2,] 16 13 NA NA NA
#> [3,] 16 13 10 NA NA
#> [4,] 16 13 10 19 NA
#> [5,] 16 13 10 19 15
Created on 2023-08-06 with reprex v2.0.2
m - 10) and the ease of referring to row/column as [1,1], etc. To be able to handle both numeric and character values, data frame (and tibble) have to resort to lists internally and it's easy to fall into the brackets when trying to address values by index. Everyone has gone through x[[1]][1] at one time or another.It is actually not. You have asked when to use [ and [[, which is exactly that: subsetting (and hence assigning on a subset).
This is explained in chapter 4 in the linked book above.
Then you are better of to use a list instead. You can define the list beforehand and then look through the list, what the output of each iteration was.
If you have a vector vec, it does not matter if you use [ or [[, but [[ makes it more clear that you refer to a single value. When you assign without [ or [[, you assign to a single variable, e.g. a scalar. So result is a scalar with one value, which can be accessed either by result, result[1] or result[[1]]. If you assign to a subset of result with [ or [[ you are actually expanding the vector/list (this expanding does not work with data.frames or matrices whatsoever, so be cautious there and define the matrix before).
So in the end everything comes down to "do you understand subsetting in R". Which is not the case at it seems to me.
Is it because if we deal with matrix or dataframe we must strictly allocate a value according to so called "coordinates" in it using like my_matrix[i,j] meaning pointing to row and column intersection to place a value there ? I hope you understand what I mean, apologies if it sounds convoluted.
Exactly, in a broader sense you have to define what happens to the values around the one you try to assign. E.g. in a data.frame, you can have different data types in different columns. But if you assign a value to a specific column in a row outside the existing ones, it is not clear what the values in all other columns should be. The same applies to a matrix (also there a no different data types between columns there).
A general advice: Always predefine the target object and it's dimensions before you apply a for loop which assigns values inside it. This will speed up your for loops drastically if they are complex enough that speed matters (e.g. if you store your results in a list, define the list with the necessary length to avoid allocation of space inside the loop in addition to the pure execution time and resources needed).
I have got another example with question , why does it not work ?
# create a list of vartable names
var_list <- c("var1", "var2", "var3")
# loop through variable names
for (var_name in var_list) {
# do something with the variable
print(paste("Values of", var_name, ":", get(var_name)))
}
from here:
https://www.projectpro.io/recipes/append-output-from-for-loop-dataframe-r#mcetoc_1gt3jqqee1e
Error in get(var_name) : object 'var1' not found
there is simply no var1, the example is incomplete as it assumes there was a var1,var2,var3 before this code snippet began
I would like to kindly ask you, could you please show me what the complete example would look like, in order for the code to work properly?
var1 <- "x"
var2 <- 99
var3 <- list(a=1,
b=2)
# create a list of vartable names
var_list <- c("var1",
"var2",
"var3")
# loop through variable names
for (var_name in var_list) {
# do something with the variable
print(paste("Values of", var_name, ":", get(var_name)))
}Thank you very much, it works now as intended.
I can see it now that var1, var2 and var3 should be defined beforehand in global environment, but would it be possible to define them inside a for loop like on-the-fly ?
I always thought that people who write tutorials should pay attention to the details so that everything works as it should.
for loops are useful in particular when you want to be operating simultaneously on a current elements and those ahead or behind in some way. for isolated or indepent iterations, its far more R like to utilise other approaches such as the base R apply* family of functions, or the tidyverse map_* equivalents; there are libraries such as slider that can help in yet more cases.
R is turing complete so in principle you can do everything in R that can be accomplished in a computer programming language. you can define variables on the fly (but you probably shouldnt ? )
The following code works, but I cant see why I would use it
library(tibble)
(mymetadata <- tibble(
varname = paste0("var",1:3),
value = list("x",99,list(a=1,b=2))
))
# create a list of vartable names
# loop through variable names
for (r in seq_len(nrow(mymetadata))) {
vn <- mymetadata[r,]$varname
assign(x = vn,
value = mymetadata[r,]$value[[1]])
# do something with the variable
print(paste("Values of", vn, ":", get(vn)))
}
Thanks a lot,
this is very helpful.
Hi again,
another for loop example and question:
results <- as.data.frame(c())
for(i in 1:10){
square <- i^2
results <- rbind(results, square)
}
results %>% rownames_to_column() %>% rename(iteration = rowname,
result_of_iteration = X1)
Is it possible to create those two columns "iteration" and "result_of_iteration" inside that for loop ?
Just to do it all at once, however this is not a big deal to do it afterwards with this line:
results %>% rownames_to_column() %>% dplyr::rename(iteration = rowname,
result_of_iteration = X1)
I am just curious how to use mutate and rename inside a for loop, if of course it makes sense ?
I have just tried this:
for(i in 1:10){
square <- i^2
results <- rbind(results, square)
return(results %>% rownames_to_column() %>% rename(iteration = rowname,
result_of_iteration = X1))
}
but this is not working.
here are 6 implementation; they arguably get worse as they get closer to 'for loops' with 'mutates'
# plain and direct
vec <- 1:10
data.frame(iteration=vec,
result_of_iteration=(vec)^2)
# if we have to mutate
library(tidyverse)
tibble(iteration=vec) |>
mutate(result_of_iteration=vec^2)
# if we want to get more involved with the loop
map_dfr(1:10,
\(x){data.frame(iteration=x,
result_of_iteration = x^2)})
# if we want to get more involved with the loop and
# are insisting on a mutate
map_dfr(1:10,
\(x){data.frame(iteration=x) |>
mutate(result_of_iteration = x^2)})
# using a for loop
df <- data.frame(matrix(nrow = 10,
ncol = 2))
names(df) <- c("iteration",
"result_of_iteration")
for(i in 1:10){
df[i,] <- data.frame(i,
i ^2)
}
df
# using a for loop
# insisting on a mutate
df <- data.frame(matrix(nrow = 10,
ncol = 2))
names(df) <- c("iteration",
"result_of_iteration")
for(i in 1:10){
df[i,] <- data.frame( a= i) |> mutate(
b = a ^2)
}
dfThank you very much indeed for comprehensive example Nir.
I have a lot to analyse about those 6 alternatives.
I have been learning about for loops all day today and just have found another example, which is brain twisting for loop solution.
Here we are:
library(dplyr)
library(tidyr)
df <- tibble(
category = c("Art","Technology","Finance"),
rating = c(100,95,50)
)
category_names <- df$category
for(name in category_names){
df <- df %>% mutate(!! name := +(category == name))
}
I would be very grateful if you could elaborate a bit what is happening in this line:
df <- df %>% mutate(!! name := +(category == name))
I suppose this has something to do with Non standard evaluation.
Particularly I would like to decipher this syntax:
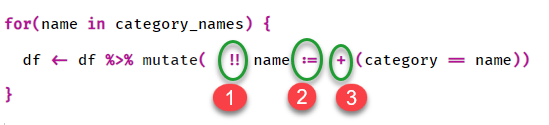
Thank you in advance.
This is from here:
https://stackoverflow.com/questions/61471128/how-to-create-columns-from-a-list-in-a-for-loop-using-mutate
the for loop causes a pass over the df 3 times, each time a column is made with the category name (!!name)
the exclamation marks (bang-bang) are to get at the contents of name rather than just be the literal text name itself. i.e. Art
when you dynamically make a column name in dplyr you are required to use the walrus operator := rather than the conventional = . for the values of the new column, category is compared to name, this gives true or false, adding a + symbol infront of TRUE/FALSE does an implicit conversoin to numeric values because addition requires numerics rather than logicals.
The loop could be replaced by the following tidyverse code
df |> mutate(dummy=1L,
cat2=category) |>
pivot_wider(names_from = "cat2",
values_from = "dummy",
values_fill = 0L)