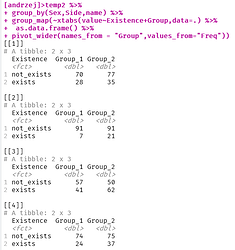
Hi,
I have got a table that consists of a few 2 by 2 tables inside it. How can I get all those tables as dataframes out of it ?
thank you
temp <- structure(list(Existence = structure(c(2L, 1L, 2L, 1L, 2L, 1L,
2L, 1L, 2L, 1L, 2L, 1L, 2L, 1L, 2L, 1L), .Label = c("not_exists",
"exists"), class = "factor"), Group = structure(c(2L, 2L, 1L,
1L, 2L, 2L, 1L, 1L, 2L, 2L, 1L, 1L, 2L, 2L, 1L, 1L), .Label = c("Group_1",
"Group_2"), class = "factor"), Sex = structure(c(1L, 1L, 1L,
1L, 1L, 1L, 1L, 1L, 2L, 2L, 2L, 2L, 2L, 2L, 2L, 2L), .Label = c("F",
"M"), class = "factor"), Side = structure(c(1L, 1L, 1L, 1L, 2L,
2L, 2L, 2L, 1L, 1L, 1L, 1L, 2L, 2L, 2L, 2L), .Label = c("right",
"left"), class = "factor"), r1 = c(35, 77, 28, 70, 37, 75, 24,
74, 23, 27, 21, 31, 20, 30, 18, 34), r2 = c(21, 91, 7, 91, 17,
95, 8, 90, 18, 32, 5, 47, 19, 31, 7, 45), r3 = c(62, 50, 41,
57, 47, 65, 35, 63, 28, 22, 29, 23, 28, 22, 31, 21)), class = "data.frame", row.names = c(NA,
-16L), variable.labels = structure(character(0), .Names = character(0)), codepage = 65001L)
(totsum1 <- sum(temp$r1+temp$r2+temp$r3))
(temp2 <- pivot_longer(data=temp,
cols=c(r1,r2,r3)))
temp3 <- temp2 %>%
uncount(weights = .$value) %>%
select(-value) %>% table()