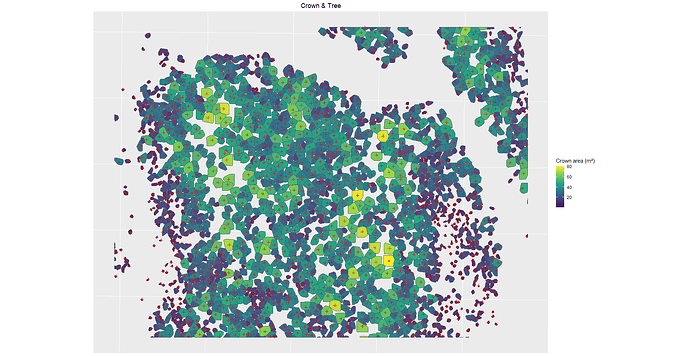
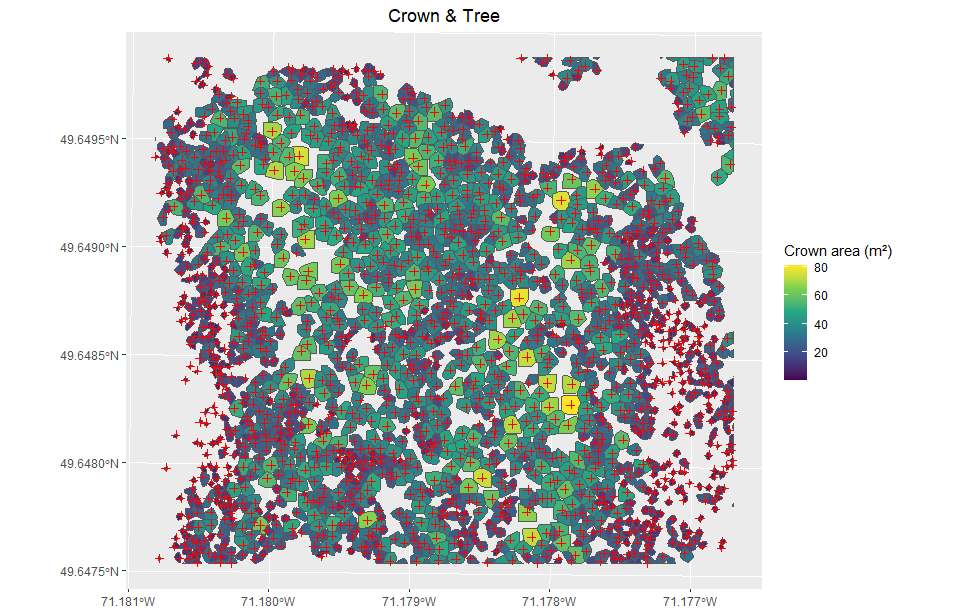
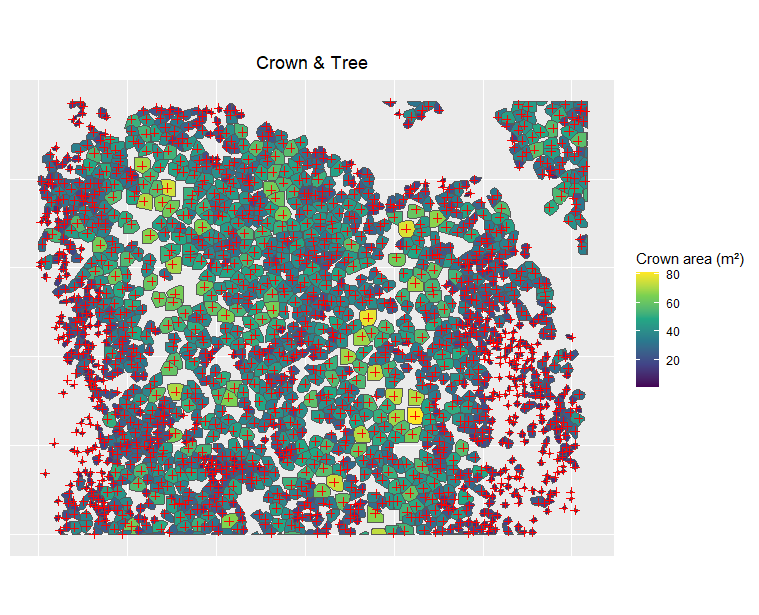
Hi, here is the result, I only did for 10 because it's a lot of information already. Let me know if you need more.
dput(head(crowns4_crop, 5))
structure(list(treeID = c(7154, 7174, 7175, 7199, 7200), Z = c(19.74775,
20.08075, 17.71525, 15.27575, 13.89975), npoints = c(473L, 394L,
311L, 432L, 323L), convhull_area = c(43.898, 33.437, 19.422,
36.496, 28.327), geometry = structure(list(structure(list(structure(c(255833.4125,
255833.23675, 255831.993001074, 255834.594286062, 255833.4125,
5501574.16775, 5501574.298, 5501575.25083612, 5501575.25083612,
5501574.16775), dim = c(5L, 2L))), class = c("XY", "POLYGON",
"sfg")), structure(list(structure(c(255856.27475, 255855.8075,
255854.6055, 255853.493676096, 255858.401473068, 255856.27475,
5501573.72825, 5501573.68075, 5501574.07975, 5501575.25083612,
5501575.25083612, 5501573.72825), dim = c(6L, 2L))), class = c("XY",
"POLYGON", "sfg")), structure(list(structure(c(255866.404, 255865.93525,
255865.393, 255863.785, 255863.205, 255863.033976154, 255866.481341375,
255866.404, 5501575.19, 5501574.92275, 5501574.61925, 5501574.72925,
5501575.09725, 5501575.25083612, 5501575.25083612, 5501575.19
), dim = c(8L, 2L))), class = c("XY", "POLYGON", "sfg")), structure(list(
structure(c(255907.4005, 255906.61925, 255906.17825, 255904.27425,
255901.62325, 255901.142, 255900.59225, 255900.586204727,
255908.506793168, 255907.4005, 5501574.0445, 5501573.728,
5501573.68025, 5501573.523, 5501574.30775, 5501574.68175,
5501575.198, 5501575.25083612, 5501575.25083612, 5501574.0445
), dim = c(10L, 2L))), class = c("XY", "POLYGON", "sfg")),
structure(list(structure(c(255935.07475, 255934.33925, 255934.28225,
255933.333063605, 255935.154217763, 255935.154217763, 255935.07475,
5501574.15, 5501574.27075, 5501574.311, 5501575.25083612,
5501575.25083612, 5501574.14661042, 5501574.15), dim = c(7L,
2L))), class = c("XY", "POLYGON", "sfg"))), class = c("sfc_POLYGON",
"sfc"), precision = 0, bbox = structure(c(xmin = 255831.993001074,
ymin = 5501573.523, xmax = 255935.154217763, ymax = 5501575.25083612
), class = "bbox"), crs = structure(list(input = "EPSG:2949",
wkt = "PROJCRS[\"NAD83(CSRS) / MTM zone 7\",\n BASEGEOGCRS[\"NAD83(CSRS)\",\n DATUM[\"NAD83 Canadian Spatial Reference System\",\n ELLIPSOID[\"GRS 1980\",6378137,298.257222101,\n LENGTHUNIT[\"metre\",1]]],\n PRIMEM[\"Greenwich\",0,\n ANGLEUNIT[\"degree\",0.0174532925199433]],\n ID[\"EPSG\",4617]],\n CONVERSION[\"MTM zone 7\",\n METHOD[\"Transverse Mercator\",\n ID[\"EPSG\",9807]],\n PARAMETER[\"Latitude of natural origin\",0,\n ANGLEUNIT[\"degree\",0.0174532925199433],\n ID[\"EPSG\",8801]],\n PARAMETER[\"Longitude of natural origin\",-70.5,\n ANGLEUNIT[\"degree\",0.0174532925199433],\n ID[\"EPSG\",8802]],\n PARAMETER[\"Scale factor at natural origin\",0.9999,\n SCALEUNIT[\"unity\",1],\n ID[\"EPSG\",8805]],\n PARAMETER[\"False easting\",304800,\n LENGTHUNIT[\"metre\",1],\n ID[\"EPSG\",8806]],\n PARAMETER[\"False northing\",0,\n LENGTHUNIT[\"metre\",1],\n ID[\"EPSG\",8807]]],\n CS[Cartesian,2],\n AXIS[\"easting (E(X))\",east,\n ORDER[1],\n LENGTHUNIT[\"metre\",1]],\n AXIS[\"northing (N(Y))\",north,\n ORDER[2],\n LENGTHUNIT[\"metre\",1]],\n USAGE[\n SCOPE[\"Engineering survey, topographic mapping.\"],\n AREA[\"Canada - Quebec - between 72°W and 69°W.\"],\n BBOX[45.01,-72,61.8,-69]],\n ID[\"EPSG\",2949]]"), class = "crs"), n_empty = 0L)), sf_column = "geometry", agr = structure(c(treeID = NA_integer_,
Z = NA_integer_, npoints = NA_integer_, convhull_area = NA_integer_
), levels = c("constant", "aggregate", "identity"), class = "factor"), row.names = c(5886L,
5900L, 5901L, 5918L, 5919L), class = c("sf", "data.table", "data.frame"
))