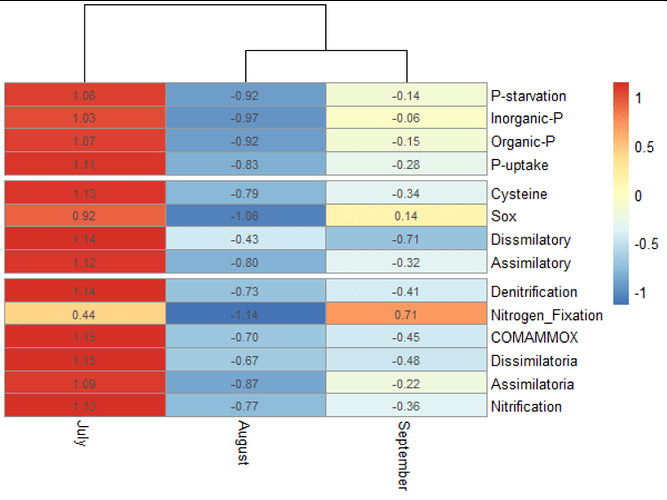
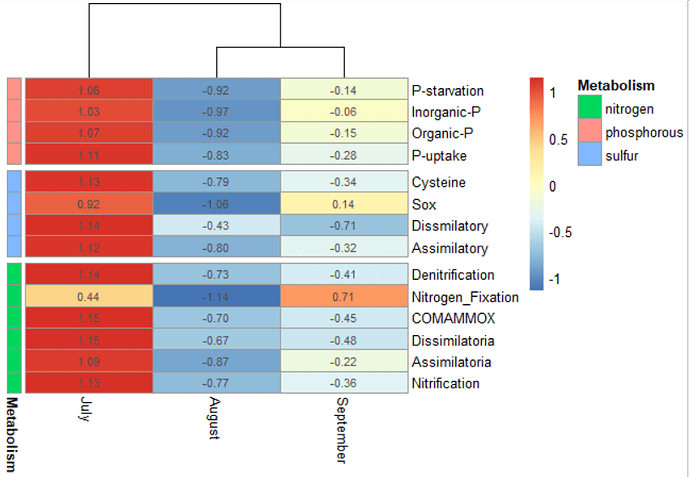
Hello everyone, I hope you are very well. I made a heatmap, which I really liked how my data is displayed. However, my data (pathways) appears out of order. Is there any way to classify certain data within the heatmap?
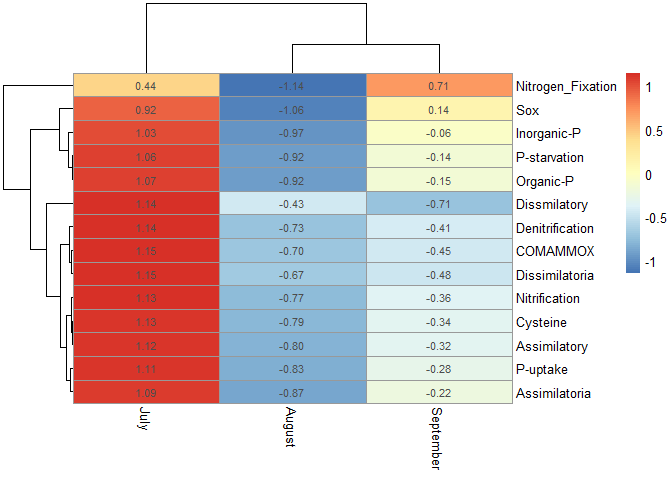
For example, P-starvation to P-uptake belong to phosphorus metabolism.
Cysteine to Assimilatory belong to sulfur metabolism, and Denitrification to Nitrification belong to nitrogen metabolism.
Could the data sort them, or could a dividing line between each metabolism?
I would like to do that in the last heatmap, blue with yellow and red.
Thanks!
# Primero creo una nueva Data a partir de tu Data original.
# Primero debes llmar a tu data original con tu script anterior.
Data <- data.frame (tibble::tribble(
~Pathways, ~July, ~August, ~September,
"P-starvation", 10462L, 7798L, 8853L,
"Inorganic-P", 4419L, 3169L, 3742L,
"Organic-P", 33916L, 24480L, 28117L,
"P-uptake", 44204L, 33561L, 36611L,
"Cysteine", 9247L, 7030L, 7551L,
"Sox", 8780L, 6351L, 7816L,
"Dissmilatory", 784L, 607L, 576L,
"Assimilatory", 30984L, 22860L, 24885L,
"Denitrification", 7303L, 5030L, 5414L,
"Nitrogen_Fixation", 561L, 411L, 587L,
"COMAMMOX", 3566L, 2455L, 2604L,
"Dissimilatoria", 8378L, 5726L, 6008L,
"Assimilatoria", 3655L, 1967L, 2524L,
"Nitrification", 144L, 107L, 115L
)
)
Data_2 <- Data
head(Data_2)
#> Pathways July August September
#> 1 P-starvation 10462 7798 8853
#> 2 Inorganic-P 4419 3169 3742
#> 3 Organic-P 33916 24480 28117
#> 4 P-uptake 44204 33561 36611
#> 5 Cysteine 9247 7030 7551
#> 6 Sox 8780 6351 7816
rownames(Data_2) <- Data_2$Pathways
Data_2 <- Data_2[-1]
class(Data_2)
#> [1] "data.frame"
MiTabla<- as.matrix(Data_2)
MiTabla
#> July August September
#> P-starvation 10462 7798 8853
#> Inorganic-P 4419 3169 3742
#> Organic-P 33916 24480 28117
#> P-uptake 44204 33561 36611
#> Cysteine 9247 7030 7551
#> Sox 8780 6351 7816
#> Dissmilatory 784 607 576
#> Assimilatory 30984 22860 24885
#> Denitrification 7303 5030 5414
#> Nitrogen_Fixation 561 411 587
#> COMAMMOX 3566 2455 2604
#> Dissimilatoria 8378 5726 6008
#> Assimilatoria 3655 1967 2524
#> Nitrification 144 107 115
MisDistancias<- dist(MiTabla, method = "euclidean")
MisDistancias
#> P-starvation Inorganic-P Organic-P P-uptake Cysteine
#> Inorganic-P 9168.8500
#> Organic-P 34633.5233 43799.2278
#> P-uptake 50722.3944 59890.6591 16138.6350
#> Cysteine 1939.3950 7261.2248 36551.6404 52634.2988
#> Sox 2449.1431 6763.2035 37048.7481 53145.0459 865.6529
#> Dissmilatory 14624.7357 5458.9949 49256.0274 65343.5822 12709.3636
#> Assimilatory 30083.9052 39248.7090 4654.7662 20658.6272 31993.0246
#> Denitrification 5428.4276 3817.9001 40024.7483 56115.5466 3513.6740
#> Nitrogen_Fixation 14863.5233 5696.0296 49495.1885 65584.7736 12952.0289
#> COMAMMOX 10730.9117 1591.3670 45355.6644 61445.1489 8813.4667
#> Dissimilatoria 4090.2647 5229.4059 38635.6033 54725.0684 2199.1876
#> Assimilatoria 10972.3312 1874.0395 45580.3131 61679.2170 9065.0517
#> Nitrification 15555.2322 6388.0042 50186.6827 66275.2724 13641.3575
#> Sox Dissmilatory Assimilatory Denitrification
#> Inorganic-P
#> Organic-P
#> P-uptake
#> Cysteine
#> Sox
#> Dissmilatory 12220.7672
#> Assimilatory 32510.2362 44700.8220
#> Denitrification 3113.8680 9244.8112 35465.7370
#> Nitrogen_Fixation 12453.6742 297.0959 44943.4015 9491.5570
#> COMAMMOX 8338.4385 3907.3536 40798.9425 5337.7799
#> Dissimilatoria 1954.7616 10647.9867 34072.6624 1411.6930
#> Assimilatoria 8572.6510 3726.5192 41029.3311 5571.5324
#> Nitrification 13148.1342 933.8742 45632.8556 10176.7682
#> Nitrogen_Fixation COMAMMOX Dissimilatoria Assimilatoria
#> Inorganic-P
#> Organic-P
#> P-uptake
#> Cysteine
#> Sox
#> Dissmilatory
#> Assimilatory
#> Denitrification
#> Nitrogen_Fixation
#> COMAMMOX 4156.4709
#> Dissimilatoria 10896.8782 6741.0682
#> Assimilatoria 3968.1155 502.4590 6969.5815
#> Nitrification 699.3490 4839.2467 11580.1281 4646.5043
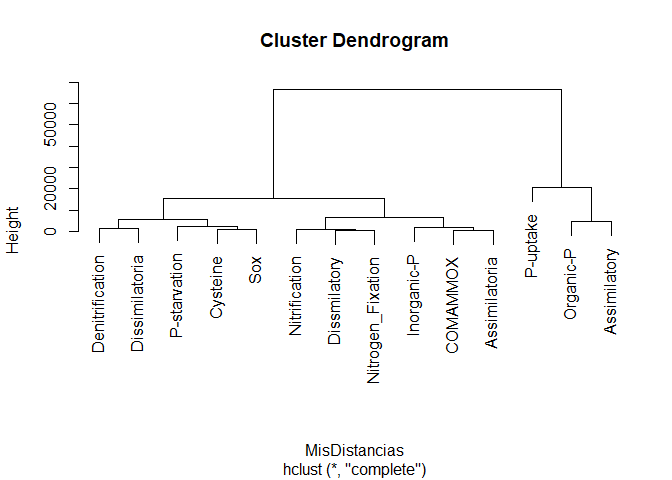
MiClusteringJerargico<- hclust(MisDistancias, method = "complete")
MiClusteringJerargico
#>
#> Call:
#> hclust(d = MisDistancias, method = "complete")
#>
#> Cluster method : complete
#> Distance : euclidean
#> Number of objects: 14
plot(MiClusteringJerargico)
MiTablaEstandarizada<- t(scale(t(MiTabla)))
# Heatmap con datos Estandarizados:
heatmap(MiTablaEstandarizada, Colv = NA, cexCol=1)
library(pheatmap)
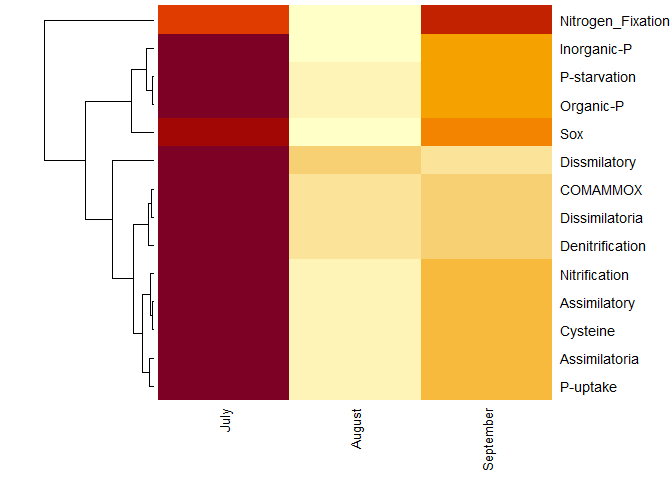
pheatmap(MiTablaEstandarizada, display_numbers = T)
Created on 2022-02-17 by the reprex package (v2.0.1)