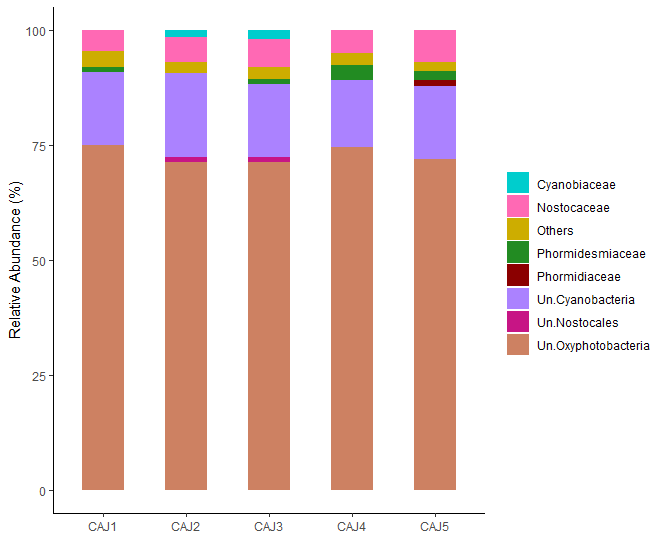
Anyone know how to set the results in my graph in ascending order?
First: Bottom : Un oxyphotobacteria, next Un cyanobacteria, nostocaceae.
My script:
library(readxl)
Family <- read_excel("~/RSTUDIO/Datos_cianobacterias/Cianobacterias_familia_sin3h5c.xlsx")
data<- Family
attach(Family)
rwnames <- index
data <- as.data.frame(data[,-1])
rownames(data) <- rwnames
Metadata<- read.csv("~/RSTUDIO/Datos_cianobacterias/Metadata-final-sin3h5c.csv", row.names=1)
#SUBSET DE SAMPLING POINTS
CAJ1 <- subset(data, Metadata$SamplingPoint == "CAJ-1", select = c(Un Caenarcaniphilales:Un Cyanobacteria))
CAJ2 <- subset(data, Metadata$SamplingPoint == "CAJ-2", select = c(Un Caenarcaniphilales:Un Cyanobacteria))
CAJ3 <- subset(data, Metadata$SamplingPoint == "CAJ-3", select = c(Un Caenarcaniphilales:Un Cyanobacteria))
CAJ4 <- subset(data, Metadata$SamplingPoint == "CAJ-4", select = c(Un Caenarcaniphilales:Un Cyanobacteria))
CAJ5 <- subset(data, Metadata$SamplingPoint == "CAJ-5", select = c(Un Caenarcaniphilales:Un Cyanobacteria))
CAJ1 <- data.frame(CAJ1)
CAJ1_counts <- colSums(CAJ1)
Counts <- unname(CAJ1_counts)
CAJ1_counts <- data.frame(CAJ1_counts)
CAJ1_counts <- t(CAJ1_counts)
total <- sum(Counts)
rel_ab <- CAJ1_counts/total
Others <- rel_ab[,colMeans(rel_ab)<.01]
Others <- sum(Others)
rel_ab <- rel_ab[,colMeans(rel_ab)>=.01]
rel_ab <- data.frame(t(rel_ab), Others)
rel_ab_P <- t(rel_ab)
abundance <- c("abundance")
rel_ab_P <- data.frame(rel_ab_P)
write.csv(rel_ab_P, file = "~/RSTUDIO/Datos_cianobacterias/CAJ1.csv")
CAJ1 <- read.csv("~/RSTUDIO/Datos_cianobacterias/CAJ1.csv")
#CAJ2
CAJ2 <- data.frame(CAJ2)
CAJ2_counts <- colSums(CAJ2)
Counts <- unname(CAJ2_counts)
CAJ2_counts <- data.frame(CAJ2_counts)
CAJ2_counts <- t(CAJ2_counts)
total <- sum(Counts)
rel_ab <- CAJ2_counts/total
Others <- rel_ab[,colMeans(rel_ab)<.01]
Others <- sum(Others)
rel_ab <- rel_ab[,colMeans(rel_ab)>=.01]
rel_ab <- data.frame(t(rel_ab), Others)
rel_ab_P <- t(rel_ab)
abundance <- c("abundance")
rel_ab_P <- data.frame(rel_ab_P)
write.csv(rel_ab_P, file = "~/RSTUDIO/Datos_cianobacterias/CAJ2.csv")
CAJ2 <- read.csv("~/RSTUDIO/Datos_cianobacterias/CAJ2.csv")
#CAJ3
CAJ3 <- data.frame(CAJ3)
CAJ3_counts <- colSums(CAJ3)
Counts <- unname(CAJ3_counts)
CAJ3_counts <- data.frame(CAJ3_counts)
CAJ3_counts <- t(CAJ3_counts)
total <- sum(Counts)
rel_ab <- CAJ3_counts/total
Others <- rel_ab[,colMeans(rel_ab)<.01]
Others <- sum(Others)
rel_ab <- rel_ab[,colMeans(rel_ab)>=.01]
rel_ab <- data.frame(t(rel_ab), Others)
rel_ab_P <- t(rel_ab)
abundance <- c("abundance")
rel_ab_P <- data.frame(rel_ab_P)
write.csv(rel_ab_P, file = "~/RSTUDIO/Datos_cianobacterias/CAJ3.csv")
CAJ3 <- read.csv("~/RSTUDIO/Datos_cianobacterias/CAJ3.csv")
#CAJ4
CAJ4 <- data.frame(CAJ4)
CAJ4_counts <- colSums(CAJ4)
Counts <- unname(CAJ4_counts)
CAJ4_counts <- data.frame(CAJ4_counts)
CAJ4_counts <- t(CAJ4_counts)
total <- sum(Counts)
rel_ab <- CAJ4_counts/total
Others <- rel_ab[,colMeans(rel_ab)<.01]
Others <- sum(Others)
rel_ab <- rel_ab[,colMeans(rel_ab)>=.01]
rel_ab <- data.frame(t(rel_ab), Others)
rel_ab_P <- t(rel_ab)
abundance <- c("abundance")
rel_ab_P <- data.frame(rel_ab_P)
write.csv(rel_ab_P, file = "~/RSTUDIO/Datos_cianobacterias/CAJ4.csv")
CAJ4 <- read.csv("~/RSTUDIO/Datos_cianobacterias/CAJ4.csv")
#CAJ5
CAJ5 <- data.frame(CAJ5)
CAJ5_counts <- colSums(CAJ5)
Counts <- unname(CAJ5_counts)
CAJ5_counts <- data.frame(CAJ5_counts)
CAJ5_counts <- t(CAJ5_counts)
total <- sum(Counts)
rel_ab <- CAJ5_counts/total
Others <- rel_ab[,colMeans(rel_ab)<.01]
Others <- sum(Others)
rel_ab <- rel_ab[,colMeans(rel_ab)>=.01]
rel_ab <- data.frame(t(rel_ab), Others)
rel_ab_P <- t(rel_ab)
abundance <- c("abundance")
rel_ab_P <- data.frame(rel_ab_P)
write.csv(rel_ab_P, file = "~/RSTUDIO/Datos_cianobacterias/CAJ5.csv")
CAJ5 <- read.csv("~/RSTUDIO/Datos_cianobacterias/CAJ5.csv")
getPalette = colorRampPalette(c("cyan3","hotpink","gold3","forestgreen","darkred","mediumpurple1","mediumvioletred","lightsalmon3","gray58","coral1","deepskyblue1","springgreen3","orchid1","yellow4"))
palette2 <- getPalette(14)
library(ggplot2)
ggplot() +geom_bar(aes(y = rel_ab_P100, x= "CAJ1", fill = X), data = CAJ1,
stat="identity", width = .5)+ geom_bar(aes(y = rel_ab_P100, x= "CAJ2", fill = X), data = CAJ2,
stat="identity",width=.5)+
geom_bar(aes(y = rel_ab_P100, x= "CAJ3", fill = X), data = CAJ3,
stat="identity", width = .5)+
geom_bar(aes(y = rel_ab_P100, x= "CAJ4", fill = X), data = CAJ4,
stat="identity", width = .5)+
geom_bar(aes(y = rel_ab_P*100, x= "CAJ5", fill = X), data = CAJ5,
stat="identity", width = .5)+
theme_classic()+
theme(legend.title = element_blank())+
ylab("Relative Abundance (%)")+
xlab(" ")+
scale_fill_manual(values = palette2)