Hi,
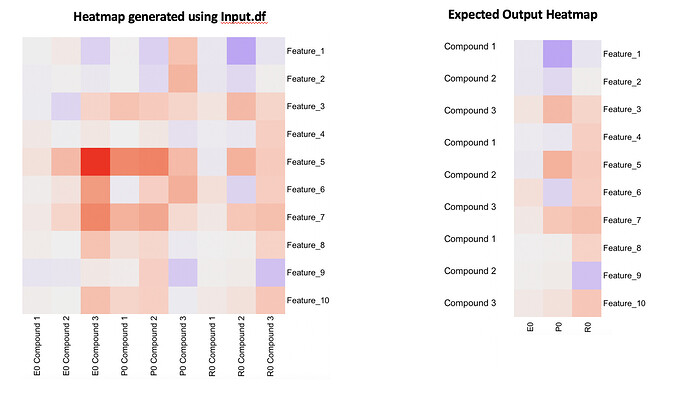
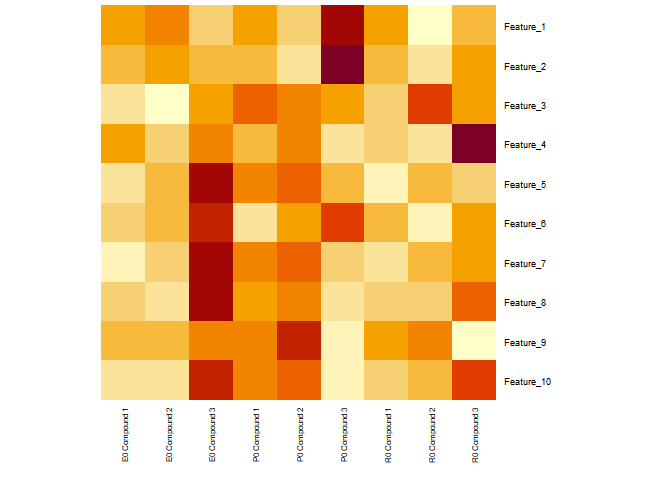
I am working with the a data matrix (features on rows and samples/compounds on columns) containing volunteers treated with different compounds. I usually use heatmap for visualisation for plotting features on y-axis (right) and samples on x-axis. This time I am interested in plotting a similar heatmap but with additional annotation on y-axis (left) (I have provided an example dataset along with the example). Basically, this will help in identifying top responsive volunteers/samples and features; that is different between volunteers for comparison.
Perhaps, I was thinking if the data matrix I am using should be reshaped or formatted before plotting. This way I can directly use this matrix heatmap. Would this be accomplished by using reshape or dplyr or tidyverse package.
dput(Input.df)
structure(list(`E0 Compound 1` = c(0.028643405, -0.170597892, -0.133431392,
0.362602406, 0.65949239, 0.129862499, 0.308021483, 0.191411185,
-0.408706956, 0.148431726), `E0 Compound 2` = c(0.312575401, 0.08023047,
-0.952555759, -0.087364689, 2.492343411, 0.594530215, 1.257403638,
-0.005561728, -0.372470805, 0.019060103), `E0 Compound 3` = c(-1.088143295,
-0.282639558, 1.283875318, 0.436371912, 7.875801388, 3.888644506,
4.851154755, 2.112242014, 0.35468163, 2.231527721), `P0 Compound 1` = c(0.0487268,
-0.00328775, 2.07844595, -0.014618298, 4.735091392, -0.220327528,
2.872283004, 0.77093127, 0.168573042, 1.237777012), `P0 Compound 2` = c(-1.125845192,
-0.801624332, 1.669743718, 0.415204306, 4.986942824, 1.496006014,
3.336380665, 1.162817817, 1.491530314, 1.484104864), `P0 Compound 3` = c(2.082646448,
2.708726878, 1.256777456, -0.491426149, 2.459378121, 2.981105751,
0.974149318, -0.222540059, -1.407150779, -0.147072039), `R0 Compound 1` = c(-0.326900367,
-0.362728527, 0.509587017, -0.165210608, -0.283082038, 0.753676032,
0.360670113, 0.037390082, 0.103973162, 0.311012902), `R0 Compound 2` = c(-2.793220245,
-0.826729075, 2.509040525, -0.344938067, 2.868355513, -1.025542457,
1.875640888, 0.096367013, 0.233463623, 0.646241163), `R0 Compound 3` = c(-0.415009733,
0.089591797, 1.245990797, 1.529969694, 1.696672088, 1.578365108,
2.218401723, 1.391261115, -1.716869822, 1.869097396)), class = "data.frame", row.names = c("Feature_1",
"Feature_2", "Feature_3", "Feature_4", "Feature_5", "Feature_6", "Feature_7", "Feature_8",
"Feature_9", "Feature_10"))
#> E0 Compound 1 E0 Compound 2 E0 Compound 3 P0 Compound 1
#> Feature_1 0.02864341 0.312575401 -1.0881433 0.04872680
#> Feature_2 -0.17059789 0.080230470 -0.2826396 -0.00328775
#> Feature_3 -0.13343139 -0.952555759 1.2838753 2.07844595
#> Feature_4 0.36260241 -0.087364689 0.4363719 -0.01461830
#> Feature_5 0.65949239 2.492343411 7.8758014 4.73509139
#> Feature_6 0.12986250 0.594530215 3.8886445 -0.22032753
#> Feature_7 0.30802148 1.257403638 4.8511548 2.87228300
#> Feature_8 0.19141119 -0.005561728 2.1122420 0.77093127
#> Feature_9 -0.40870696 -0.372470805 0.3546816 0.16857304
#> Feature_10 0.14843173 0.019060103 2.2315277 1.23777701
#> P0 Compound 2 P0 Compound 3 R0 Compound 1 R0 Compound 2
#> Feature_1 -1.1258452 2.0826464 -0.32690037 -2.79322025
#> Feature_2 -0.8016243 2.7087269 -0.36272853 -0.82672908
#> Feature_3 1.6697437 1.2567775 0.50958702 2.50904053
#> Feature_4 0.4152043 -0.4914261 -0.16521061 -0.34493807
#> Feature_5 4.9869428 2.4593781 -0.28308204 2.86835551
#> Feature_6 1.4960060 2.9811058 0.75367603 -1.02554246
#> Feature_7 3.3363807 0.9741493 0.36067011 1.87564089
#> Feature_8 1.1628178 -0.2225401 0.03739008 0.09636701
#> Feature_9 1.4915303 -1.4071508 0.10397316 0.23346362
#> Feature_10 1.4841049 -0.1470720 0.31101290 0.64624116
#> R0 Compound 3
#> Feature_1 -0.4150097
#> Feature_2 0.0895918
#> Feature_3 1.2459908
#> Feature_4 1.5299697
#> Feature_5 1.6966721
#> Feature_6 1.5783651
#> Feature_7 2.2184017
#> Feature_8 1.3912611
#> Feature_9 -1.7168698
#> Feature_10 1.8690974
dput(Sample_metadata.df)
structure(list(Volunteer = c("E0", "E0", "E0", "P0", "P0", "P0",
"R0", "R0", "R0"), Compounds = c("Compound 1", "Compound 2", "Compound 3", "Compound 1",
"Compound 2", "Compound 3", "Compound 1", "Compound 2", "Compound 3")), class = "data.frame", row.names = c("E0 Compound 1",
"E0 Compound 2", "E0 Compound 3", "P0 Compound 1", "P0 Compound 2", "P0 Compound 3",
"R0 Compound 1", "R0 Compound 2", "R0 Compound 3"))
#> Volunteer Compounds
#> E0 Compound 1 E0 Compound 1
#> E0 Compound 2 E0 Compound 2
#> E0 Compound 3 E0 Compound 3
#> P0 Compound 1 P0 Compound 1
#> P0 Compound 2 P0 Compound 2
#> P0 Compound 3 P0 Compound 3
#> R0 Compound 1 R0 Compound 1
#> R0 Compound 2 R0 Compound 2
#> R0 Compound 3 R0 Compound 3
Created on 2022-06-03 by the reprex package (v2.0.1)
Thank you,
Toufiq