Hi,
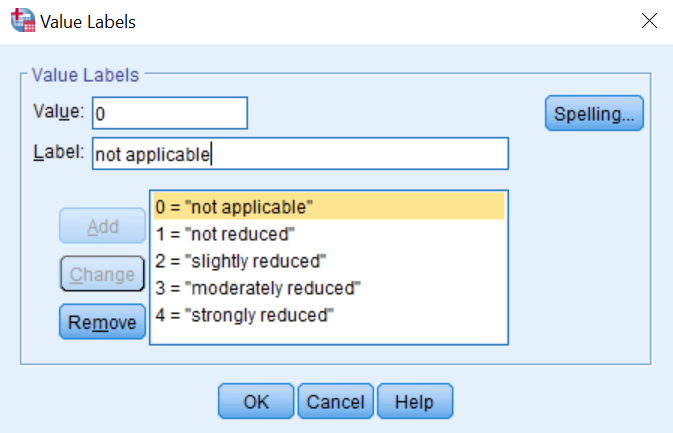
I would like to recode a few variables in order for them to have the same Likert scale coding.
I have done it easily in SPSS but somehow I can't replicate it in R.
So I have two questions:
- Maybe is there a package to transfer spss files to R with recoded values left intact ?
- How to allocate these Likert scale re-coding to many (d1:d9) variables in R at once ?
I feel that I should use mutate_at and across functions, but would be grateful for hint how to start.
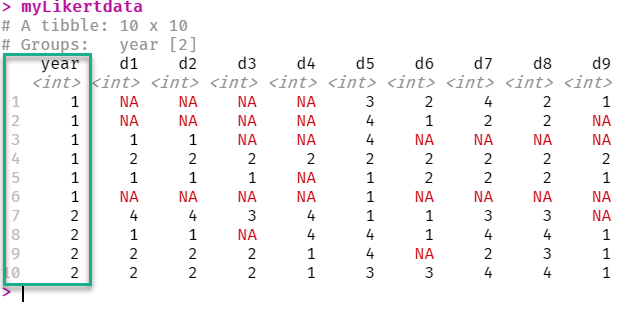
myLikertdata <- structure(list(year = structure(c(1L, 1L, 1L, 1L, 1L, 1L, 2L,
2L, 2L, 2L), .Label = c("pre-clinical", "clinical"), class = "factor"),
d1 = c(NA, NA, 1L, 2L, 1L, NA, 4L, 1L, 2L, 2L), d2 = c(NA,
NA, 1L, 2L, 1L, NA, 4L, 1L, 2L, 2L), d3 = c(NA, NA, NA, 2L,
1L, NA, 3L, NA, 2L, 2L), d4 = c(NA, NA, NA, 2L, NA, NA, 4L,
4L, 1L, 1L), d5 = c(3L, 4L, 4L, 2L, 1L, 1L, 1L, 4L, 4L, 3L
), d6 = c(2L, 1L, NA, 2L, 2L, NA, 1L, 1L, NA, 3L), d7 = c(4L,
2L, NA, 2L, 2L, NA, 3L, 4L, 2L, 4L), d8 = c(2L, 2L, NA, 2L,
2L, NA, 3L, 4L, 3L, 4L), d9 = c(1L, NA, NA, 2L, 1L, NA, NA,
1L, 1L, 1L)), class = c("grouped_df", "tbl_df", "tbl", "data.frame"
), row.names = c(NA, -10L), groups = structure(list(year = structure(1:2, .Label = c("pre-clinical",
"clinical"), class = "factor"), .rows = structure(list(1:6, 7:10), ptype = integer(0), class = c("vctrs_list_of",
"vctrs_vctr", "list"))), class = c("tbl_df", "tbl", "data.frame"
), row.names = c(NA, -2L), .drop = TRUE))
myLikertdata$year <- factor(myLikertdata$year, levels=c('1','2'), labels=c('pre-clinical','clinical'))
Likert_scale <- c("not reduced" = 1, "slightly reduced" = 2, "
moderately reduced" = 3, "strongly reduced" = 4)
and I would like to apply this Likert_scale to all variables from d1 to d9.
Any help would be much appreciated.