I am a fan of the marginaleffects package and really appreciate its concise and easy-to-understand syntax. Whenever I encounter the emmeans function, I prefer to convert it into marginaleffects to study it. However, I recently came across this code, and I’m stuck—I can’t figure out how to proceed. The problem is as follows:
library(tidyverse)
library(lme4)
library(emmeans)
library(marginaleffects)
d <- mtcars %>%
as_tibble() %>%
select(cyl, vs, am, gear, carb) %>%
mutate(cyl = cyl > 6)
m <- lme4::glmer(
cyl ~ vs * am + (1 | gear / carb),
data = d,
family = binomial
)
m %>%
emmeans::emmeans(specs = ~ vs + am, type = "response") %>%
pairs(infer = T, adjust = "tukey") %>%
as.data.frame()
I can recode first line
m %>%
emmeans::emmeans(specs = ~ vs + am, type = "response") %>%
as
library(marginaleffects)
m %>%
marginaleffects::predictions(
newdata = datagrid(vs = unique, am = unique),
type = "response",
re.form = NA
) %>%
as_tibble()
but I can’t figure out the second line.
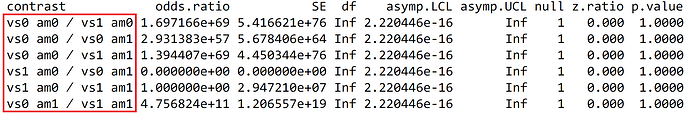
m %>%
emmeans::emmeans(specs = ~ vs + am, type = "response") %>%
pairs(infer = T, adjust = "tukey") #<<
Could you please give me some help? Thank you so much!