Hi,
I have a gene array in form of a matrix [y, x] where y is a vector of outcome and x is a data frame with the predictors as columns. It is a matrix because it is to be used in glmnet() for ridge and lasso. However, before I use it (glmnet(y~x)…), I need to split it randomly 50/50 into training and testing datasets. How do I do that?
Many thanks!
Suppose the matrix is called m. You can choose rows with 50/50 chance of being chosen by
inTraining <- sample(c(0,1), size = nrow(m), replace = TRUE)
training <- m(inTraining, )
testing <- m(1-inTraining, )
My matrix named 'data' looks like this:
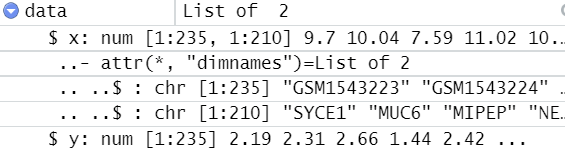
@startz, when I put size = nrow(data), I get an error that size argument is invalid.
I wonder whether R knows it's a matrix. Try nrow(as.matrix(data))
The best thing that worked so far was to put x in nrow(x) since x is the data frame part. However, now y is too long for the models. Any advice is much appreciated.
That's helpful. I see now that data is not a matrix, it is a list of a matrix and a vector. Try something like
inTraining <- sample(c(0,1), size = nrow(data$x), replace = TRUE)
training <- list(x = data$x[inTraining, ], y = data$y[inTraining])
testing <- list(x = data$x[1 - inTraining, ], y = data$y[1 - inTraining])
Thank you so much! The code you suggested worked. One more thing - do I need to add set.seed(1) before the code to make this partition reproducible?
Yes. Doesn't have to be 1. All that matters is that set.seed() always gets the same argument.
This topic was automatically closed 7 days after the last reply. New replies are no longer allowed.
If you have a query related to it or one of the replies, start a new topic and refer back with a link.