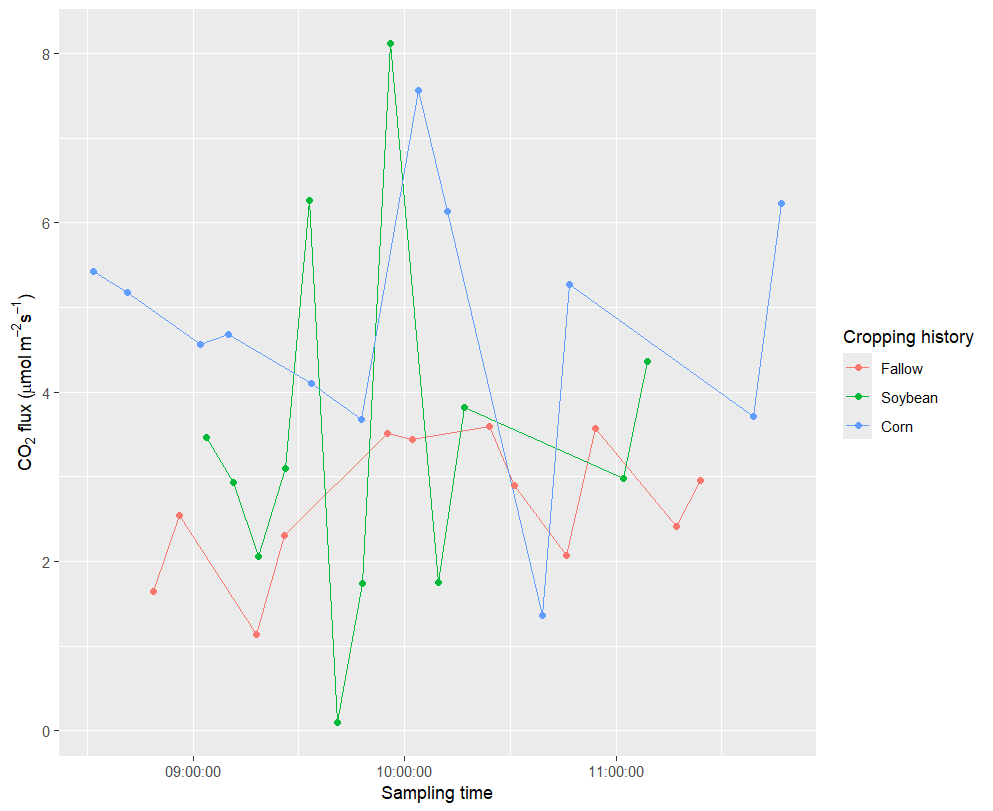
Please considering this plot, where I am using the hms package to convert the time to actual time. how can I show all the time point in this plot using ggplot2 ?. Also I need the errorbar to appear in each point?
Thank you very much for the help.
library(tidyverse)
library(hms)
Data
dat2 <- structure(
list(
Cropping history = structure(
c(
3L,
3L,
1L,
1L,
2L,
2L,
2L,
2L,
3L,
3L,
1L,
1L,
2L,
2L,
1L,
1L,
3L,
3L,
3L,
3L,
1L,
1L,
2L,
2L,
2L,
2L,
3L,
3L,
1L,
1L,
2L,
2L,
1L,
1L,
3L,
3L
),
levels = c("Fallow", "Soybean", "Corn"),
class = "factor"
),
[YYYY-MM-DD HH:MM:SS] = structure(
c(
1746435720,
1746436200,
1746436680,
1746437160,
1746437580,
1746438060,
1746438480,
1746438960,
1746439440,
1746439920,
1746441960,
1746442440,
1746442920,
1746443340,
1746443820,
1746444240,
1746445140,
1746445620,
1747297902,
1747298477,
1747298923,
1747299366,
1747299827,
1747300281,
1747300719,
1747301164,
1747301609,
1747302462,
1747302910,
1747303338,
1747303780,
1747304220,
1747304649,
1747305075,
1747305538,
1747305999
),
class = c("POSIXct", "POSIXt"),
tzone = "UTC"
),
FCO2_DRY LIN = c(
4.56358,
4.68132,
1.14151,
2.30355,
6.26688,
0.09713,
1.73805,
8.12184,
7.56254,
6.1318,
2.07687,
3.57559,
2.97575,
4.36482,
2.41366,
2.96188,
3.71283,
6.22674,
5.42002,
5.17549,
1.64929,
2.54588,
3.46512,
2.93409, 2.05559, 3.09399, 4.09809, 3.6724, 3.51691, 3.44157,
1.75552, 3.81452, 3.59866, 2.9023, 1.36774, 5.27381)), row.names = c(NA,
-36L), class = c("tbl_df", "tbl", "data.frame"))
ghg$date <- lubridate::ymd_hms(ghg$[YYYY-MM-DD HH:MM:SS])
ggplot(data = ghg,aes(x = as_hms(date),y = FCO2_DRY LIN,colour=Cropping history,group=Cropping history))+
stat_summary(geom = 'line',fun = mean)+
stat_summary(geom = 'point',fun = mean)+
stat_summary(geom = 'errorbar',fun.data = mean_se)+
#scale_x_time()+
labs(x='Sampling time', y = expression(paste(CO[2], " flux (", mu, "mol "*m^{-2}*s^{-1}, ")")))