Dear R users,
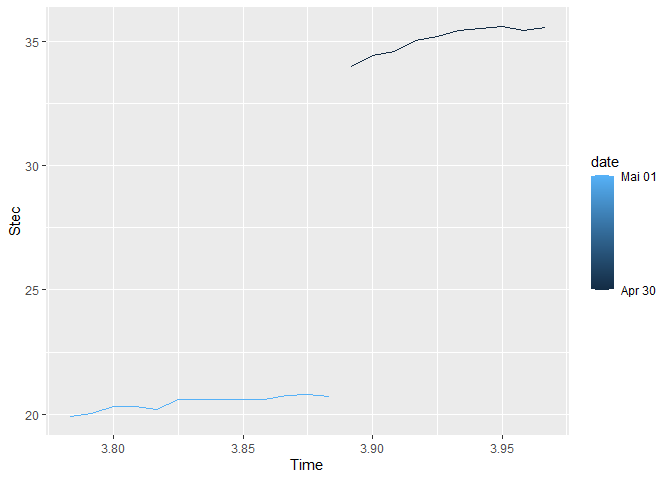
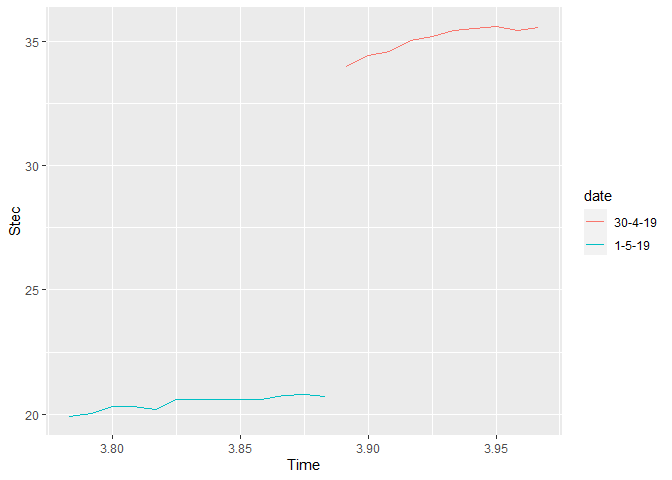
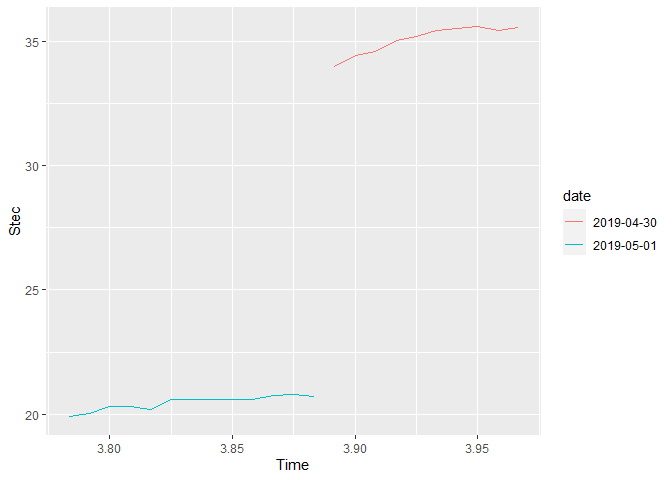
I am trying to create a multiple time-series plot using R and trying to create legend based on the dates. I have obtained the graph but it doesn't look so neat and the legends are not in proper order.
The script which I used is,
library(ggplot2)
library(readxl)
D1 <- read_excel("E:/Gokul/R 1.xlsx",
sheet = "30-4-19")
D2 <- read_excel("E:/Gokul/R 1.xlsx",
sheet = "1-5-19")
D3 <- read_excel("E:/Gokul/R 1.xlsx",
sheet = "2-5-19")
D4 <- read_excel("E:/Gokul/R 1.xlsx",
sheet = "3-5-19")
D5<- read_excel("E:/Gokul/R 1.xlsx",
sheet = "4-5-19")
D6 <- read_excel("E:/Gokul/R 1.xlsx",
sheet = "5-5-19")
D7 <- read_excel("E:/Gokul/R 1.xlsx",
sheet = "6-5-19")
D8 <- read_excel("E:/Gokul/R 1.xlsx",
sheet = "7-5-19")
D9 <- read_excel("E:/Gokul/R 1.xlsx",
sheet = "8-5-19")
D10 <- read_excel("E:/Gokul/R 1.xlsx",
sheet = "9-5-19")
D11 <- read_excel("E:/Gokul/R 1.xlsx",
sheet = "10-5-19")
D12 <- read_excel("E:/Gokul/R 1.xlsx",
sheet = "11-5-19")
D13 <- read_excel("E:/Gokul/R 1.xlsx",
sheet = "12-5-19")
D14 <- read_excel("E:/Gokul/R 1.xlsx",
sheet = "13-5-19")
D15 <- read_excel("E:/Gokul/R 1.xlsx",
sheet = "14-5-19")
ggplot(D1,aes(x=Time,y=Stec))+
geom_line(aes(color="30-4-19"))+
geom_line(data=D2,aes(color="1-5-19"))+
geom_line(data=D3,aes(color="2-5-19"))+
geom_line(data=D4,aes(color="3-5-19"))+
geom_line(data=D5,aes(color="4-5-19"))+
geom_line(data=D6,aes(color="5-5-19"))+
geom_line(data=D7,aes(color="6-5-19"))+
geom_line(data=D8,aes(color="7-5-19"))+
geom_line(data=D9,aes(color="8-5-19"))+
geom_line(data=D10,aes(color="9-5-19"))+
geom_line(data=D11,aes(color="10-5-19"))+
geom_line(data=D12,aes(color="11-5-19"))+
geom_line(data=D13,aes(color="12-5-19"))+
geom_line(data=D14,aes(color="13-5-19"))+
geom_line(data=D15,aes(color="14-5-19"))+
labs(color="Legend text")
The graph which I got is
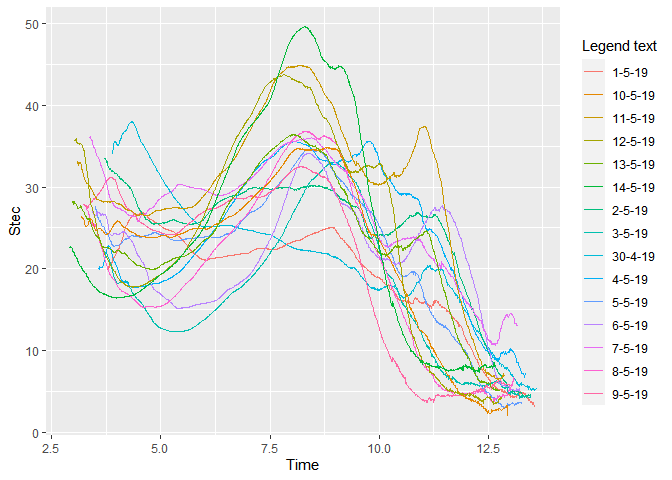
How do I make the graph more appealing, interpretable and make the legends in ascending order.?
Kindly suggest the solution for the above problem
Thank you.