Dear all!
I hope you are healthy.
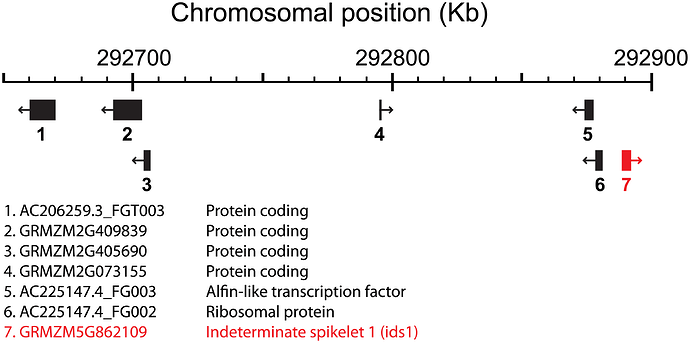
I have a data set like this
gene chr start end
yala 1A 677309370 677309490
yaba 1A 335767676 335728901
yaca 1A 587656732 589037382
yada 1A 604098765 604284729
yaea 1A 169376234 169409823
I am planning to visualize the data like this
I need your help to suggest me what kind of packages that I could use to make the data in X-axis with position "start" and "end" based on my data?
Please help me. It is really urgents.
I really thank for kind people.
Best