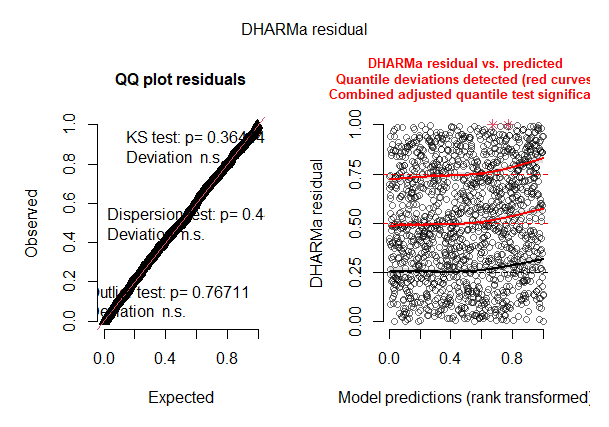
I'm having trouble understanding what it means to have excessively large C.I. on estimates of variance. I'm using a generalized additive mixed effect model (GAMM), but maybe this effect applies to GLMs as well. All other diagnostics (via package DHARMa) say the model is fine: Pearson correlation between predictor variables is < 0.47, no zero-inflation, no over-dispersion, qq-plot lines up well, no glaring issues in summary output, and seemingly no strong spatial or temporal auto-correlation.
So, based on the results below from mgcv::gam.vcomp, is this model showing signs of being overfit, or is it just having trouble estimating the C.I. of something with almost no effect. As in, nothing to worry about (?)
library(dplyr)
library(mgcv)
library(DHARMa)
toad3 <- subset(toad2, fSite %in% c(1,10,20,30,40),
select = c(CYR, fCYR, fSeason, fSite, area_sampled, num))
dput(toad3)
structure(list(CYR = c(2008L, 2008L, 2008L, 2008L, 2008L, 2008L,
2008L, 2008L, 2009L, 2009L, 2009L, 2009L, 2009L, 2009L, 2009L,
2009L, 2009L, 2009L, 2010L, 2010L, 2010L, 2010L, 2010L, 2010L,
2010L, 2010L, 2010L, 2011L, 2011L, 2011L, 2011L, 2011L, 2011L,
2011L, 2011L, 2011L, 2011L, 2012L, 2012L, 2012L, 2012L, 2012L,
2012L, 2012L, 2012L, 2012L, 2012L, 2013L, 2013L, 2013L, 2013L,
2013L, 2013L, 2013L, 2013L, 2013L, 2013L, 2014L, 2014L, 2014L,
2014L, 2014L, 2014L, 2014L, 2014L, 2014L, 2015L, 2015L, 2015L,
2015L, 2015L, 2015L, 2015L, 2015L, 2015L, 2015L, 2016L, 2016L,
2016L, 2016L, 2016L, 2016L, 2016L, 2016L, 2016L, 2016L, 2017L,
2017L, 2017L, 2017L, 2017L, 2017L, 2017L, 2017L, 2017L, 2018L,
2018L, 2018L, 2018L, 2018L, 2018L, 2018L, 2018L, 2018L, 2018L,
2019L, 2019L, 2019L, 2019L, 2019L, 2019L, 2019L, 2019L, 2019L,
2019L, 2020L, 2020L, 2020L, 2020L, 2020L, 2020L, 2020L, 2020L,
2020L, 2020L, 2021L, 2021L, 2021L, 2021L, 2021L, 2022L, 2022L,
2022L, 2022L, 2022L, 2022L, 2022L, 2022L, 2022L, 2023L, 2023L,
2023L, 2023L, 2023L, 2023L, 2023L, 2023L, 2023L, 2023L, 2024L,
2024L, 2024L, 2024L, 2024L), fCYR = structure(c(1L, 1L, 1L, 1L,
1L, 1L, 1L, 1L, 2L, 2L, 2L, 2L, 2L, 2L, 2L, 2L, 2L, 2L, 3L, 3L,
3L, 3L, 3L, 3L, 3L, 3L, 3L, 4L, 4L, 4L, 4L, 4L, 4L, 4L, 4L, 4L,
4L, 5L, 5L, 5L, 5L, 5L, 5L, 5L, 5L, 5L, 5L, 6L, 6L, 6L, 6L, 6L,
6L, 6L, 6L, 6L, 6L, 7L, 7L, 7L, 7L, 7L, 7L, 7L, 7L, 7L, 8L, 8L,
8L, 8L, 8L, 8L, 8L, 8L, 8L, 8L, 9L, 9L, 9L, 9L, 9L, 9L, 9L, 9L,
9L, 9L, 10L, 10L, 10L, 10L, 10L, 10L, 10L, 10L, 10L, 11L, 11L,
11L, 11L, 11L, 11L, 11L, 11L, 11L, 11L, 12L, 12L, 12L, 12L, 12L,
12L, 12L, 12L, 12L, 12L, 13L, 13L, 13L, 13L, 13L, 13L, 13L, 13L,
13L, 13L, 14L, 14L, 14L, 14L, 14L, 15L, 15L, 15L, 15L, 15L, 15L,
15L, 15L, 15L, 16L, 16L, 16L, 16L, 16L, 16L, 16L, 16L, 16L, 16L,
17L, 17L, 17L, 17L, 17L), levels = c("2008", "2009", "2010",
"2011", "2012", "2013", "2014", "2015", "2016", "2017", "2018",
"2019", "2020", "2021", "2022", "2023", "2024"), class = "factor"),
fSeason = structure(c(1L, 1L, 1L, 2L, 2L, 2L, 2L, 2L, 1L,
1L, 1L, 1L, 1L, 2L, 2L, 2L, 2L, 2L, 1L, 1L, 1L, 1L, 2L, 2L,
2L, 2L, 2L, 1L, 1L, 1L, 1L, 1L, 2L, 2L, 2L, 2L, 2L, 1L, 1L,
1L, 1L, 1L, 2L, 2L, 2L, 2L, 2L, 1L, 1L, 1L, 1L, 1L, 2L, 2L,
2L, 2L, 2L, 1L, 1L, 1L, 1L, 2L, 2L, 2L, 2L, 2L, 1L, 1L, 1L,
1L, 1L, 2L, 2L, 2L, 2L, 2L, 1L, 1L, 1L, 1L, 1L, 2L, 2L, 2L,
2L, 2L, 2L, 2L, 2L, 2L, 2L, 1L, 1L, 1L, 1L, 1L, 1L, 1L, 1L,
1L, 2L, 2L, 2L, 2L, 2L, 1L, 1L, 1L, 1L, 1L, 2L, 2L, 2L, 2L,
2L, 1L, 1L, 1L, 1L, 1L, 2L, 2L, 2L, 2L, 2L, 2L, 2L, 2L, 2L,
2L, 1L, 1L, 1L, 1L, 1L, 2L, 2L, 2L, 2L, 1L, 1L, 1L, 1L, 1L,
2L, 2L, 2L, 2L, 2L, 1L, 1L, 1L, 1L, 1L), levels = c("DRY",
"WET"), class = "factor"), fSite = structure(c(10L, 1L, 20L,
40L, 20L, 30L, 10L, 1L, 40L, 30L, 20L, 10L, 1L, 10L, 1L,
20L, 30L, 40L, 40L, 30L, 20L, 1L, 1L, 10L, 40L, 30L, 20L,
30L, 20L, 10L, 1L, 40L, 40L, 30L, 20L, 1L, 10L, 40L, 30L,
20L, 1L, 10L, 40L, 20L, 30L, 10L, 1L, 40L, 30L, 20L, 10L,
1L, 40L, 30L, 20L, 10L, 1L, 30L, 20L, 10L, 1L, 40L, 30L,
20L, 1L, 10L, 40L, 30L, 20L, 10L, 1L, 40L, 20L, 30L, 10L,
1L, 40L, 30L, 20L, 10L, 1L, 40L, 30L, 20L, 10L, 1L, 1L, 10L,
40L, 30L, 20L, 40L, 20L, 10L, 1L, 40L, 30L, 20L, 10L, 1L,
10L, 1L, 40L, 30L, 20L, 10L, 1L, 40L, 30L, 20L, 40L, 30L,
1L, 20L, 10L, 40L, 30L, 20L, 10L, 1L, 20L, 1L, 10L, 40L,
30L, 20L, 30L, 40L, 10L, 1L, 40L, 1L, 10L, 20L, 30L, 1L,
10L, 40L, 30L, 10L, 40L, 20L, 30L, 1L, 40L, 20L, 30L, 1L,
10L, 30L, 1L, 10L, 40L, 20L), levels = c("1", "2", "3", "4",
"5", "6", "7", "8", "9", "10", "11", "12", "13", "14", "15",
"16", "17", "18", "19", "20", "21", "22", "23", "24", "25",
"26", "27", "28", "29", "30", "31", "32", "33", "34", "35",
"36", "37", "38", "39", "40", "41", "42", "43", "44", "45",
"46", "47"), class = "factor"), area_sampled = c(3L, 3L,
3L, 3L, 3L, 3L, 3L, 3L, 3L, 3L, 3L, 3L, 3L, 3L, 3L, 3L, 3L,
3L, 3L, 3L, 3L, 3L, 3L, 3L, 3L, 3L, 3L, 3L, 3L, 3L, 3L, 3L,
3L, 3L, 3L, 3L, 3L, 3L, 3L, 3L, 3L, 3L, 3L, 3L, 3L, 3L, 3L,
3L, 3L, 3L, 3L, 3L, 3L, 3L, 3L, 3L, 3L, 3L, 3L, 3L, 3L, 3L,
3L, 3L, 3L, 3L, 3L, 3L, 3L, 3L, 3L, 3L, 3L, 3L, 3L, 3L, 3L,
3L, 3L, 3L, 3L, 3L, 3L, 3L, 3L, 3L, 3L, 3L, 3L, 3L, 3L, 3L,
3L, 3L, 3L, 3L, 3L, 3L, 3L, 3L, 3L, 3L, 3L, 3L, 3L, 3L, 3L,
3L, 3L, 3L, 3L, 3L, 3L, 3L, 3L, 3L, 3L, 3L, 3L, 3L, 3L, 3L,
3L, 3L, 3L, 3L, 3L, 3L, 3L, 3L, 3L, 3L, 3L, 3L, 3L, 3L, 3L,
3L, 3L, 3L, 3L, 3L, 3L, 3L, 3L, 3L, 3L, 3L, 3L, 3L, 3L, 3L,
3L, 3L), num = c(0L, 0L, 0L, 0L, 1L, 0L, 5L, 0L, 0L, 0L,
0L, 0L, 0L, 1L, 0L, 0L, 0L, 6L, 1L, 0L, 0L, 0L, 0L, 5L, 0L,
0L, 1L, 0L, 1L, 0L, 0L, 0L, 6L, 0L, 0L, 1L, 1L, 0L, 0L, 0L,
0L, 0L, 0L, 1L, 0L, 0L, 0L, 0L, 4L, 0L, 2L, 0L, 1L, 0L, 1L,
0L, 2L, 0L, 0L, 0L, 0L, 0L, 0L, 0L, 0L, 0L, 0L, 0L, 0L, 0L,
0L, 0L, 0L, 0L, 0L, 0L, 0L, 0L, 1L, 0L, 1L, 0L, 0L, 0L, 0L,
0L, 0L, 0L, 1L, 1L, 0L, 2L, 0L, 3L, 0L, 3L, 0L, 0L, 0L, 0L,
1L, 0L, 1L, 0L, 0L, 0L, 0L, 0L, 1L, 0L, 4L, 0L, 0L, 0L, 0L,
2L, 0L, 0L, 0L, 0L, 0L, 0L, 0L, 1L, 5L, 0L, 1L, 0L, 1L, 0L,
0L, 0L, 0L, 0L, 0L, 0L, 0L, 1L, 0L, 0L, 4L, 0L, 1L, 0L, 0L,
0L, 1L, 0L, 0L, 2L, 1L, 0L, 0L, 0L)), row.names = c(1L, 10L,
20L, 22L, 31L, 41L, 42L, 52L, 70L, 78L, 86L, 104L, 110L, 115L,
126L, 134L, 144L, 154L, 166L, 172L, 185L, 190L, 196L, 197L, 213L,
227L, 239L, 248L, 258L, 263L, 271L, 281L, 296L, 309L, 319L, 323L,
324L, 342L, 352L, 362L, 369L, 377L, 384L, 400L, 410L, 418L, 424L,
432L, 444L, 456L, 462L, 470L, 477L, 486L, 500L, 509L, 517L, 526L,
539L, 546L, 554L, 563L, 574L, 585L, 592L, 593L, 618L, 628L, 632L,
634L, 642L, 653L, 670L, 680L, 681L, 691L, 709L, 713L, 728L, 732L,
739L, 745L, 759L, 769L, 773L, 780L, 798L, 799L, 816L, 819L, 829L,
855L, 857L, 865L, 874L, 887L, 894L, 904L, 912L, 921L, 929L, 938L,
949L, 961L, 970L, 976L, 982L, 989L, 999L, 1011L, 1024L, 1046L,
1047L, 1057L, 1061L, 1072L, 1081L, 1090L, 1099L, 1109L, 1125L,
1128L, 1136L, 1139L, 1160L, 1166L, 1176L, 1186L, 1194L, 1204L,
1212L, 1220L, 1227L, 1239L, 1249L, 1258L, 1259L, 1269L, 1278L,
1296L, 1303L, 1313L, 1323L, 1335L, 1345L, 1355L, 1366L, 1374L,
1375L, 1393L, 1403L, 1412L, 1420L, 1430L), class = "data.frame")
# fit model
mod <- gam(num ~ # Abundance
# Long-term annual trend
s(CYR) +
# Seasonal variation of annual trend
s(CYR, by = fSeason) +
fSeason +
# Repeated measure / random intercept: "Not all sites have the same abundance"
s(fSite, bs = "re") +
# Random slopes per site: "Sites have their own annual trend"
s(fSite, CYR, bs = "re") +
offset(log(area_sampled)), # Area sampled offset term
data = toad2,
method = 'REML',
select = FALSE,
gamma = 1.4,
# 1.4 can be a sensible choice for suppressing over-fitting - S. Wood
family = nb(link = "log"),
control = list(trace = TRUE),
drop.unused.levels=FALSE)
> mgcv::gam.vcomp(mod)
Standard deviations and 0.95 confidence intervals:
std.dev lower upper
s(CYR) 0.0056061735 1.153835e-101 2.723888e+96
s(CYR):fSeasonDRY 0.0671182937 7.620192e-03 5.911748e-01
s(CYR):fSeasonWET 0.7120572644 2.750020e-01 1.843716e+00
s(fSite) 0.0182263177 5.112121e-94 6.498255e+89
s(fSite,CYR) 0.0004233687 3.039279e-04 5.897486e-04
Rank: 5/5
> summary(mod)
Family: Negative Binomial(0.329)
Link function: log
Formula:
num ~ s(CYR) + s(CYR, by = fSeason) + fSeason + s(fSite, bs = "re") +
s(fSite, CYR, bs = "re") + offset(log(area_sampled))
Parametric coefficients:
Estimate Std. Error z value Pr(>|z|)
(Intercept) -3.2675 0.1834 -17.815 < 2e-16 ***
fSeasonWET 0.5919 0.1793 3.301 0.000965 ***
---
Signif. codes: 0 ‘***’ 0.001 ‘**’ 0.01 ‘*’ 0.05 ‘.’ 0.1 ‘ ’ 1
Approximate significance of smooth terms:
edf Ref.df Chi.sq p-value
s(CYR) 1.00795 1.012 70.696 <2e-16 ***
s(CYR):fSeasonDRY 1.09186 1.605 110.166 <2e-16 ***
s(CYR):fSeasonWET 6.42196 7.460 76.441 <2e-16 ***
s(fSite) 0.01465 46.000 0.018 0.135
s(fSite,CYR) 32.10291 46.000 147.831 <2e-16 ***
---
Signif. codes: 0 ‘***’ 0.001 ‘**’ 0.01 ‘*’ 0.05 ‘.’ 0.1 ‘ ’ 1
Rank: 122/123
R-sq.(adj) = 0.158 Deviance explained = 39.4%
-REML = 629.91 Scale est. = 1 n = 1436
library(DHARMa)
simulationOutput <- simulateResiduals(fittedModel = mod)
plot(simulationOutput)