I have this data set here:
library(igraph)
data = structure(list(Adenocalymma_adenophorum = c(NA, 0, 0, 0, 0.003122927,
0.00999241, 0.008685473, 0.007730365, 0, 0, 0, 0, 0, 0, 0, 0,
0, 0.003573423, 0, 0, 0, 0, 0), Adenocalymma_cf_bracteosum = c(0L,
NA, 0L, 0L, 0L, 0L, 0L, 0L, 0L, 0L, 0L, 0L, 0L, 0L, 0L, 0L, 0L,
0L, 0L, 0L, 0L, 0L, 0L), Adenocalymma_flaviflorum = c(0L, 0L,
NA, 0L, 0L, 0L, 0L, 0L, 0L, 0L, 0L, 0L, 0L, 0L, 0L, 0L, 0L, 0L,
0L, 0L, 0L, 0L, 0L), Adenocalymma_japurensis = c(0L, 0L, 0L,
NA, 0L, 0L, 0L, 0L, 0L, 0L, 0L, 0L, 0L, 0L, 0L, 0L, 0L, 0L, 0L,
0L, 0L, 0L, 0L), Adenocalymma_longilinium = c(0.18893711, 0,
0, 0, NA, 0.183237263, 0.139293056, 0.120902907, 0, 0, 0, 0,
0, 0, 0, 0, 0, 0, 0, 0.132071652, 0, 0.457142857, 0.114500717
), Adenocalymma_moringifolium = c(0.322255215, 0, 0, 0, 0.097676062,
NA, 0.261095249, 0.131416203, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0.146191646,
0, 0, 0, 0), Adenocalymma_neoflavidum = c(0.086854728, 0, 0,
0, 0.023023646, 0.080959767, NA, 0.034786642, 0, 0, 0, 0, 0,
0, 0, 0, 0, 0, 0, 0, 0, 0, 0), Adenocalymma_tanaeciicarpum = c(0.09469697,
0, 0, 0, 0.024480341, 0.049917782, 0.042613636, NA, 0, 0, 0,
0, 0, 0, 0, 0, 0, 0, 0.255554962, 0, 0, 0, 0), Amphilophium_parkeri = c(0L,
0L, 0L, 0L, 0L, 0L, 0L, 0L, NA, 0L, 0L, 0L, 0L, 0L, 0L, 0L, 0L,
0L, 0L, 0L, 0L, 0L, 0L), Anemopaegma_robustum = c(0L, 0L, 0L,
0L, 0L, 0L, 0L, 0L, 0L, NA, 0L, 0L, 0L, 0L, 0L, 0L, 0L, 0L, 0L,
0L, 0L, 0L, 0L), Bignonia_aequinoctialis = c(0L, 0L, 0L, 0L,
0L, 0L, 0L, 0L, 0L, 0L, NA, 0L, 0L, 0L, 0L, 0L, 0L, 0L, 0L, 0L,
0L, 0L, 0L), Bignonia_prieurei = c(0L, 0L, 0L, 0L, 0L, 0L, 0L,
0L, 0L, 0L, 0L, NA, 0L, 0L, 0L, 0L, 0L, 0L, 0L, 0L, 0L, 0L, 0L
), Calichlamys_latifolia = c(0L, 0L, 0L, 0L, 0L, 0L, 0L, 0L,
0L, 0L, 0L, 0L, NA, 0L, 0L, 0L, 0L, 0L, 0L, 0L, 0L, 0L, 0L),
Fridericia_cf_trailli = c(0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0.765625,
0, 0, NA, 0, 0, 0, 0, 0, 0, 0, 0, 0), Fridericia_chica = c(0L,
0L, 0L, 0L, 0L, 0L, 0L, 0L, 0L, 0L, 0L, 0L, 0L, 0L, NA, 0L,
0L, 0L, 0L, 0L, 0L, 0L, 0L), Fridericia_cinamomea = c(0L,
0L, 0L, 0L, 0L, 0L, 0L, 0L, 0L, 0L, 0L, 0L, 0L, 0L, 0L, NA,
0L, 0L, 0L, 0L, 0L, 0L, 0L), Fridericia_nigrescens = c(0L,
0L, 0L, 0L, 0L, 0L, 0L, 0L, 0L, 0L, 0L, 0L, 0L, 0L, 0L, 0L,
NA, 0L, 0L, 0L, 0L, 0L, 0L), Fridericia_prancei = c(0.040201005,
0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, NA, 0, 0,
0, 0, 0), Fridericia_triplinervia = c(0, 0, 0, 0, 0, 0.041930937,
0, 0.192970073, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, NA, 0, 0, 0,
0), Pachyptera_aromatica = c(0, 0, 0, 0, 0.030562035, 0,
0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, NA, 0, 0, 0), Pleonotoma_albiflora = c(0L,
0L, 0L, 0L, 0L, 0L, 0L, 0L, 0L, 0L, 0L, 0L, 0L, 0L, 0L, 0L,
0L, 0L, 0L, 0L, NA, 0L, 0L), Pleonotoma_melioides = c(0,
0, 0, 0, 0.151121606, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0,
0, 0, 0, 0, NA, 0.039751553), Tynanthus_panurensis = c(0,
0, 0, 0, 0, 0.026693325, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0,
0, 0, 0, 0, 0.011428571, NA)), class = "data.frame", row.names = c("Adenocalymma_adenophorum",
"Adenocalymma_cf_bracteosum", "Adenocalymma_flaviflorum", "Adenocalymma_japurensis",
"Adenocalymma_longilinium", "Adenocalymma_moringifolium", "Adenocalymma_neoflavidum",
"Adenocalymma_tanaeciicarpum", "Amphilophium_parkeri", "Anemopaegma_robustum",
"Bignonia_aequinoctialis", "Bignonia_prieurei", "Calichlamys_latifolia",
"Fridericia_cf_trailli", "Fridericia_chica", "Fridericia_cinamomea",
"Fridericia_nigrescens", "Fridericia_prancei", "Fridericia_triplinervia",
"Pachyptera_aromatica", "Pleonotoma_albiflora", "Pleonotoma_melioides",
"Tynanthus_panurensis"))
View (data)
class (data)
data= data.matrix(data, rownames.force = NA) #
class (data)
network <- graph_from_adjacency_matrix (data)
Graph construction:
library(ggraph)
ggraph(graph,layout = "circle")+
geom_node_point(shape=19, size=3)+
geom_node_text(aes(size =40, label = name),family="serif")+
geom_edge_link0(aes(edge_width = weight),edge_colour = "black",
arrow = arrow(angle = 30, length = unit(2, "mm"),
ends = "last", type = "closed"))+
scale_fill_manual()+
scale_edge_width(range = c(0.2,2))+
scale_size(range = c(1,10))+
theme_graph()+
theme(legend.position = "none")
I tried to make my graph look like this one:
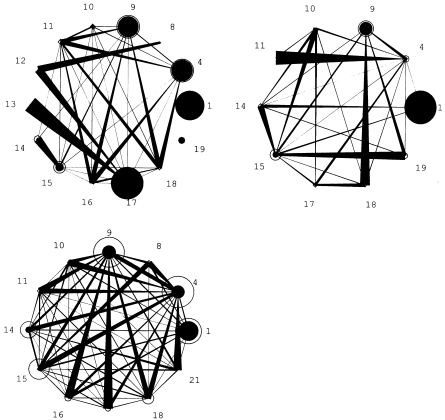
However, I’ve been trying for a few hours, but I haven’t been able to:
- increase the nodes according to their values (I managed to do this using the igraph, but not using the ggraph, I migrated to the ggraph because I was not able to change the width of the edges according to their values by the igraph);
- Modify the location of the label vertex so that it is visible;
Thank you in advance to anyone who can help me. If you know how to change the width of the edges according to their values through the igraph, it can also be. I think my chart is a little bit more advanced by igraph.
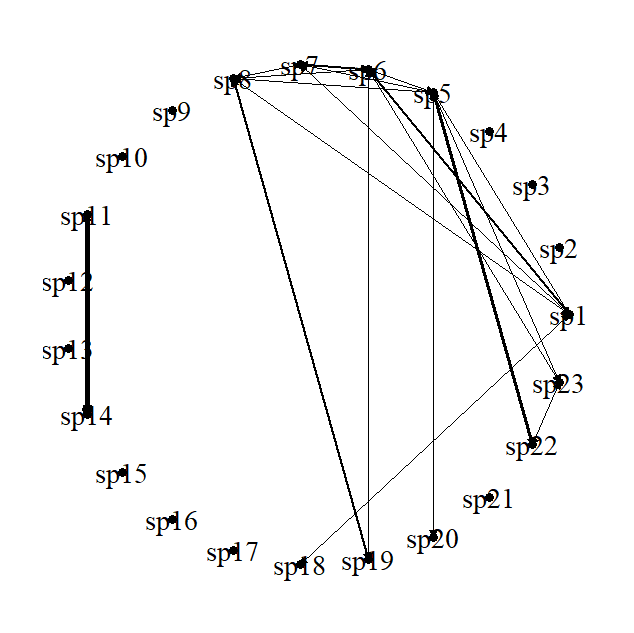


 . May I ask you to please check this question for being solved?
. May I ask you to please check this question for being solved?