The dataset contains key variables (year, month and country), and the predictors are many (in the real dataset).
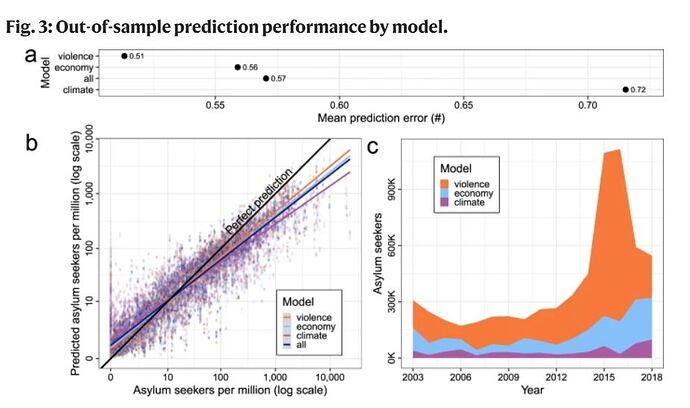
Let´s say that a and b are part of the group of climate variables and c and d the violent[enter image description here][1] group. The aim is to get metrics by "climate" and "violent"
I want to get the RMSE by a group of variables but not by each variable. Still, I am keeping getting results by variable...
Source of the picture: article
countries <- data.frame(
expand.grid(country = c("Angola", "South Sudan", "Namibia"), year = 2006:2019, month = 1:12),
deaths = round(runif(9, 1000, 20000), 0),
a = c(6, 7, 4),
b = c(5, 8, 9),
c = c(2, 20, 80),
d = c(100, 300, 500)
)
#Let´s say that "a" and "b" are part of the group of climate variables
# and "c" and "d" the violent group
#My Key ids are country, year and month
##Open libraries
library(tidymodels)
library(parsnip)
library(forcats)
library(ranger)
library(baguette)
library(lubridate)
library(ranger)
library(DALEX)
library(rlang)
library(future)
###########################################################
set.seed(123)
#split this single dataset into two: a training set and a testing set
data_split <- initial_split(countries)
# Create data frames for the two sets:
train_data <- training(data_split)
test_data <- testing(data_split)
# resample the data with 10-fold cross-validation (10-fold by default)
cv <- vfold_cv(train_data, v=3)
###########################################################
##Produce the recipe
#getting errors with "rec"
rec <- recipe(deaths~ ., data = countries) %>%
step_nzv(all_predictors(), freq_cut = 0, unique_cut = 0) %>% # remove variables with zero variances
step_novel(all_nominal()) %>% # prepares test data to handle previously unseen factor levels
step_impute_median()(all_numeric(), -all_outcomes(), -has_role("id vars")) %>% # replaces missing numeric observations with the median
step_dummy(all_nominal(), -has_role("id vars")) # dummy codes categorical variables
#OR?
my_recipe <- train_data%>%
recipe(deaths~ .) %>%
add_role(a,b, new_role = "climate") %>%
add_role(c, d, new_role = "conflict") %>%
#step_rm(iso3c, year, month) %>%
#step_dummy(all_nominal()) %>%
step_novel(all_nominal()) %>%
step_impute_median((all_numeric())) %>%
step_string2factor(all_nominal())%>%
prep()
my_recipe
###################################################################################
###################################################
##Random forests
###################################################
mod_rf <-rand_forest(trees = 1e3) %>%
set_engine("ranger",
num.threads = parallel::detectCores(),
importance = "permutation",
verbose = TRUE) %>%
set_mode("regression")
##Create Workflow
wflow_rf <- workflow() %>%
add_model(mod_rf) %>%
add_recipe(my_recipe)
##Fit the model
plan(multisession)
fit_rf<-fit_resamples(
wflow_rf,
cv,
metrics = metric_set(rmse, rsq),
control = control_resamples(save_pred = TRUE,
extract = function(x) extract_model(x)))
# extract roots
rf_tree_roots <- function(x){
map_chr(1:1000,
~ranger::treeInfo(x, tree = .)[1, "splitvarName"])
}
rf_roots <- function(x){
x %>%
select(.extracts) %>%
unnest(cols = c(.extracts)) %>%
mutate(oob_rmse = map_dbl(.extracts,
~sqrt(.x$prediction.error)),
roots = map(.extracts,
~rf_tree_roots(.))
) %>%
dplyr::select(roots) %>%
unnest(cols = c(roots))
}
#
# plot
rf_roots(fit_rf) %>%
group_by(roots) %>%
count() %>%
dplyr::arrange(desc(n)) %>%
dplyr::filter(n > 75) %>%
ggplot(aes(fct_reorder(roots, n), n)) +
geom_col() +
coord_flip() +
labs(x = "root", y = "count")