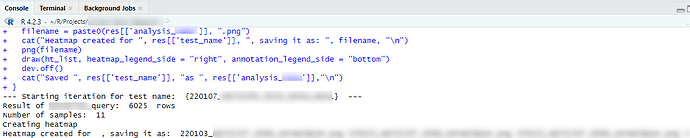
Hello, I am trying to create separate heatmap images in r. The below script will create only one image. The 'test_name' in the script has multiple tests and I would like to create separate images for each test. What adjustments should I make in the script to make this possible?
'sql1' is my connection string and 'query' is the query used to pull data from DB
test_name = '{TEST_NAME}'
res = dbGetQuery(sql1, gsub("\\{TEST_NAME\\}", test_name, gsub("\\n\\s+", " ", query)))
res[['status']] = ifelse(res[['line']] == 'TRUE', 'N', 'T')
res[is.na(res[['spec_name']]),][['spec_name']] = res[is.na(res[['spec_name']]),][['number']]
res[['spec_desc']] = paste0(res[['last_name']], '__', res[['spec_name']])
variants = unique(res[['hash']])
samples = unique(res[['spec_desc']])
dat = res[,c('hash','spec_desc', 'varfreq')] %>%
group_by(hash,spec_desc) %>%
summarize(mean_varfreq = mean(varfreq))
df = data.frame(variant = variants)
for (sample in samples) {
df = merge(df, dat[dat[['sample_desc']] == sample, c('hash','mean_varfreq')], by.x=c('variant'), by.y=c('hash'), all.x=TRUE)
names(df)[names(df) == 'mean_varfreq'] = sample
}
rownames(df) = df[['variant']]
df_binary = df
df_binary[['variant']] = NULL
df_binary[is.na(df_binary)] = 0
df_binary = df_binary[apply(df_binary, 1, max, na.rm=TRUE) >= 40,]
df_binary[df_binary > 0] = 1
cormat = cor(df_binary)
cormat = cormat[order(rownames(cormat)), order(colnames(cormat))]
h_map = Heatmap(as.matrix(cormat), #dat_row_scaled,
name = "spec info",
col = colorRamp2(c(0,0.5,1), c(colors[2], "white", colors[1])),
cluster_columns=TRUE,
cluster_rows=TRUE,
show_column_dend=FALSE,
show_row_dend=FALSE,
show_column_names = TRUE,
show_row_names = TRUE,
column_names_gp = gpar(fontsize = font_size * 0.8),
row_names_gp = gpar(fontsize = font_size * 0.8),
row_names_side = "left",
column_title = "",
column_title_gp = gpar(fontsize = font_size * 0.9),
heatmap_legend_param = list(direction = "vertical"))
ht_list = h_map
png("res[['spec_name']].png")
draw(ht_list, heatmap_legend_side = "right", annotation_legend_side = "bottom")