Hi there, I am working inside a research environment remote desktop and have built a cross-validated random forest model. I saved the workflow model as the vetiver model in a model board at the remote desktop and downloaded it from the remote desktop onto my own personal computer as a RDS file. However, I am unable to apply it in my PC to an external dataset. I would really appreciate any guidance on this.
Can you share a little more about the kind of code that is causing problems for you? If I were wanting to do this, I would start out by storing the model on the first machine with something like this:
library(tidymodels)
library(vetiver)
#>
#> Attaching package: 'vetiver'
#> The following object is masked from 'package:tune':
#>
#> load_pkgs
library(pins)
data(Sacramento)
rf_spec <- rand_forest(mode = "regression")
rf_form <- price ~ type + sqft + beds + baths
rf_fit <-
workflow(rf_form, rf_spec) %>%
fit(Sacramento)
v <- vetiver_model(rf_fit, "sacramento_rf")
model_board <- board_folder(path = "/tmp/test")
vetiver_pin_write(model_board, v)
#> Creating new version '20240925T201217Z-73f49'
#> Writing to pin 'sacramento_rf'
#>
#> Create a Model Card for your published model
#> • Model Cards provide a framework for transparent, responsible reporting
#> • Use the vetiver `.Rmd` template as a place to start
Created on 2024-09-25 with reprex v2.1.1
And then on the second machine, I would read the model and predict() like this:
library(tidymodels)
library(vetiver)
#>
#> Attaching package: 'vetiver'
#> The following object is masked from 'package:tune':
#>
#> load_pkgs
library(pins)
data(Sacramento)
model_board <- board_folder(path = "/tmp/test")
v <- vetiver_pin_read(model_board, "sacramento_rf")
predict(v, Sacramento[5:10,])
#> # A tibble: 6 × 1
#> .pred
#> <dbl>
#> 1 106683.
#> 2 133271.
#> 3 147360.
#> 4 158743.
#> 5 128204.
#> 6 155486.
Created on 2024-09-25 with reprex v2.1.1
Are you trying to do something different?
Thanks so much for your prompt reply. So I can't use the same board folder to read the model. I downloaded it in my PC. I have downloaded the "v" from the remote desktop as a .RDS file. When I tried to use readRDS and then use it for prediction, the error message I am getting is attached.
Would really appreciate help with this. Thanks again.
Data splitting
set.seed(23102013)
dta_split <- initial_split(dta1, prop = 3/5)
dta_train = training(dta_split)
dta_test = testing(dta_split)
dta_fold = mc_cv(dta_train, times = 50)
nrow(dta_train)
nrow(dta_test)
Model building
tree_rec = recipe(No_Nadir~., data = dta_train)%>%
update_role(OS, All.Cause.Mortality, new_role = "id variable")%>%
step_center(AGE,ECOGBL,PSABL,HGBBL,BASEBMI)%>%
step_scale(AGE,ECOGBL,PSABL,HGBBL,BASEBMI)%>%
step_dummy(all_nominal_predictors(), one_hot = F)
tune_spec = rand_forest(
mtry = tune(),
trees = 1000,
min_n = tune())%>%
set_mode("classification")%>%
set_engine("ranger", importance = "impurity")
tune_wf = workflow()%>%
add_model(tune_spec)%>%
add_recipe(tree_rec)
Crossvalidation and tuning hyperparameters with grid search
library(parallel)
library(doParallel)
cl <- makePSOCKcluster(2)
registerDoParallel(cl)
set.seed(23102013)
tune_res = tune_grid(
tune_wf,
resamples = dta_fold,
grid = 1000,
control = control_resamples(save_pred = T)
)
tune_res%>%
collect_metrics()%>%
filter(.metric == "roc_auc")%>%
dplyr::select(mean, min_n, mtry)%>%
pivot_longer(min_n:mtry, values_to = "value", names_to = "parameters")%>%
ggplot(aes(value,mean,color=parameters)) +
geom_point(show.legend = F) +
facet_wrap(~parameters, scales = "free_x") +
labs(x = NULL, y = "AUC")
Selecting the best model
### Selecting best parameters
best_auc <- select_best(tune_res)
final_rf = finalize_model(tune_spec,
best_auc)
model_board = board_folder("/document/psa_response").
v <- veiver_pin_read(model_board, "final_rf").
###Downloaded v from the remote environment as a RDS file.
##Now in my Personal desktop
final_model = readRDS("v.RDS")
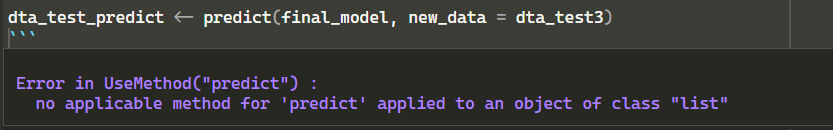
predict(final_model, dta_test3)I would really appreciate it if someone could please help me understand where I was wrong.
Could you update your example to use the reprex package and perhaps a simple dataset that we both have access to? Using reprex makes it easier to see both the input and output, and for us to re-run the code in a local session. Thanks! ![]()
Julia,
My apologies for a late reply. I got sick. Also, I somehow could not use reprex but I did run the codes with a simple dataset (iris). And I still get the error message. Here is what I am getting.
library(tidymodels)
library(tidyverse)
library(caret)
library(vetiver)
library(pins)
library(themis)
library(tidyverse)
library(reprex)
data("iris")
set.seed(123)
trees_split <- initial_split(iris, prop = 3/5)
trees_train <- training(trees_split)
trees_test <- testing(trees_split)
# How many cores does your CPU have
library(parallel)
library(doParallel)
n_cores <- parallel::detectCores()
n_cores
# Register cluster
cluster <- makeCluster(n_cores - 2)
registerDoParallel(cluster)
tree_rec <- recipe(Species ~ ., data = trees_train)
tune_spec <- rand_forest(
mtry = tune(),
trees = 250,
min_n = tune()
) %>%
set_mode("classification") %>%
set_engine("ranger")
tune_wf <- workflow() %>%
add_recipe(tree_rec) %>%
add_model(tune_spec)
set.seed(234)
trees_folds <- vfold_cv(trees_train,5)
doParallel::registerDoParallel()
set.seed(345)
tune_res <- tune_grid(
tune_wf,
resamples = trees_folds,
grid = 10
)
tune_res
##############################################################################3
best_auc <- select_best(tune_res)
final_rf = finalize_model(tune_spec,
best_auc)
final_rf_wf <- workflow()%>%
add_recipe(tree_rec)%>%
add_model(final_rf)
fitted_wf <- final_rf_wf%>%
fit(trees_train)
v <- vetiver_model(fitted_wf, "final_rf")
model_board = board_folder("C:/Users/soumy/Documents/For Upload")
vetiver_pin_write(model_board, v)
###Downloaded v from the remote environment as a RDS file.
##Now in my Personal desktop
final_rf <- readRDS("~/For Upload/final_rf/20241011T163532Z-df082/final_rf.rds")
predict(final_rf, trees_test)
Instead of this:
final_rf <- readRDS("~/For Upload/final_rf/20241011T163532Z-df082/final_rf.rds")
predict(final_rf, trees_test)
Try this:
model_board <- board_folder(path = "C:/Users/soumy/Documents/For Upload")
final_rf <- vetiver_pin_read(model_board, "final_rf")
predict(final_rf, trees_test)
This topic was automatically closed 90 days after the last reply. New replies are no longer allowed.
If you have a query related to it or one of the replies, start a new topic and refer back with a link.