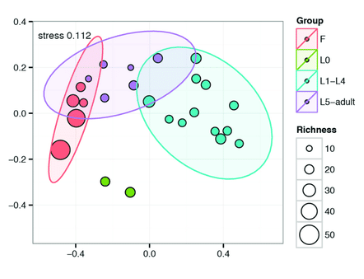
taken from the article:

taken from the article:
Hi @Manu2592. Can you provide some sample data, so we can show you how to do this.
Thanks for the reply, I attached an example of the data.
It shows the abundance of the species found in samples at 4 monitoring points during 3 different months.
The nMDS wishes to do so using the Bray Curtis similarity index
@Manu2592. You can use metaMDS function from vegan package and get ordination by scores.
library(tidyverse)
library(vegan)
#> Loading required package: permute
#> Loading required package: lattice
#> This is vegan 2.5-6
df <- strsplit("Group Estacion Richness Especie1 Especie2 Especie3 Especie4 Especie5 Especie6 Especie7
Agosto E1 6 87 87 89 91 87 94 0
Agosto E2 7 77 78 78 77 95 45 45
Agosto E3 7 85 87 89 89 78 89 95
Agosto E4 3 57 56 54 0 0 0 0
Diciembre E1 6 77 78 78 77 95 45 0
Diciembre E2 7 65 64 68 69 65 64 69
Diciembre E3 7 74 71 75 75 76 81 75
Diciembre E4 2 38 39 0 0 0 0 0
Abril E1 7 81 82 79 82 78 79 82
Abril E2 7 69 71 72 71 68 69 73
Abril E3 7 74 79 78 75 79 75 76
Abril E4 3 51 52 49 0 0 0 0", "\n") %>%
unlist() %>%
strsplit("\t") %>%
unlist() %>%
matrix(ncol = 10, byrow = TRUE) %>%
{`colnames<-`(data.frame(.[-1,], stringsAsFactors = FALSE), .[1,])} %>%
mutate_at(vars(-Group, -Estacion), as.numeric)
nmds <- metaMDS(select(df, starts_with("Especie")))
#> Square root transformation
#> Wisconsin double standardization
#> Run 0 stress 9.50621e-05
#> Run 1 stress 9.28732e-05
#> ... New best solution
#> ... Procrustes: rmse 0.07745138 max resid 0.1635964
#> Run 2 stress 8.426199e-05
#> ... New best solution
#> ... Procrustes: rmse 0.04876778 max resid 0.1076306
#> Run 3 stress 9.264371e-05
#> ... Procrustes: rmse 0.1273014 max resid 0.3039066
#> Run 4 stress 9.982599e-05
#> ... Procrustes: rmse 0.1460939 max resid 0.3519883
#> Run 5 stress 9.321337e-05
#> ... Procrustes: rmse 0.1530958 max resid 0.307541
#> Run 6 stress 9.841593e-05
#> ... Procrustes: rmse 0.1452649 max resid 0.2305473
#> Run 7 stress 8.436978e-05
#> ... Procrustes: rmse 0.100593 max resid 0.1857994
#> Run 8 stress 0.1170156
#> Run 9 stress 9.526842e-05
#> ... Procrustes: rmse 0.1228197 max resid 0.2700057
#> Run 10 stress 9.027838e-05
#> ... Procrustes: rmse 0.04196788 max resid 0.07499517
#> Run 11 stress 9.205092e-05
#> ... Procrustes: rmse 0.1711949 max resid 0.3601161
#> Run 12 stress 9.491741e-05
#> ... Procrustes: rmse 0.166596 max resid 0.309751
#> Run 13 stress 9.60547e-05
#> ... Procrustes: rmse 0.1493274 max resid 0.2815726
#> Run 14 stress 8.786096e-05
#> ... Procrustes: rmse 0.1360596 max resid 0.2699309
#> Run 15 stress 0.3238549
#> Run 16 stress 9.492202e-05
#> ... Procrustes: rmse 0.04303402 max resid 0.07046762
#> Run 17 stress 0.1170156
#> Run 18 stress 9.509689e-05
#> ... Procrustes: rmse 0.08498762 max resid 0.1882996
#> Run 19 stress 9.248478e-05
#> ... Procrustes: rmse 0.1545526 max resid 0.3351295
#> Run 20 stress 9.882223e-05
#> ... Procrustes: rmse 0.06788452 max resid 0.1553638
#> *** No convergence -- monoMDS stopping criteria:
#> 17: stress < smin
#> 3: stress ratio > sratmax
#> Warning in metaMDS(select(df, starts_with("Especie"))): stress is (nearly) zero:
#> you may have insufficient data
scores(nmds) %>%
cbind(df) %>%
ggplot(aes(x = NMDS1, y = NMDS2)) +
geom_point(aes(size = Richness, color = Group)) +
stat_ellipse(geom = "polygon", aes(group = Group, color = Group, fill = Group), alpha = 0.3) +
annotate("text", x = -2, y = 0.95, label = paste0("stress: ", format(nmds$stress, digits = 4)), hjust = 0) +
theme_bw()
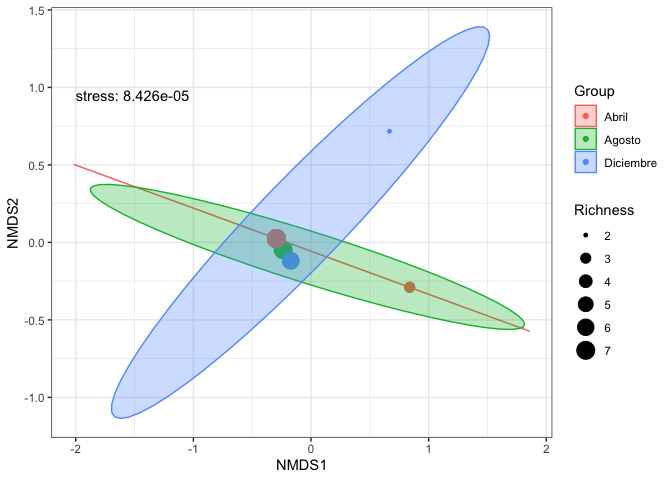
Created on 2020-01-02 by the reprex package (v0.3.0)
Many thanks for the answer, is what I was looking for.
And if I don't want to do quadratic transformation but logarithmic, could it be?
@Manu2592. You can log transform the data before preforming NMDS.
This topic was automatically closed 21 days after the last reply. New replies are no longer allowed.