> compound_name <- c("2-Hydroxybutyric acid","1-Methyladenosine","tt","2-Aminooctanoic acid")
> name2refmet(compound_name)
trying URL 'https://www.metabolomicsworkbench.org/rest/refmet/match/2-Hydroxybutyricacid/name/txt'
Error in utils::download.file(url, ref) :
cannot open URL 'https://www.metabolomicsworkbench.org/rest/refmet/match/2-Hydroxybutyricacid/name/txt'
In addition: Warning messages:
1: In file.remove(ref) :
cannot remove file 'test.data.1', reason 'No such file or directory'
2: In utils::download.file(url, ref) :
URL ' ': status was 'SSL connect error'
> > .libPaths()
Error: unexpected '>' in ">"
When I apply this code block it always replies me with the above error, I tried to handle it but failed. Please help me how to handle this error.
You'll need to do this.
library(jsonlite)
item <- fromJSON('https://www.metabolomicsworkbench.org/rest/refmet/match/2-Hydroxybutyricacid/name/txt')
item
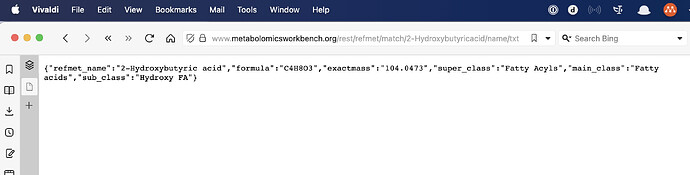
#> $refmet_name
#> [1] "2-Hydroxybutyric acid"
#>
#> $formula
#> [1] "C4H8O3"
#>
#> $exactmass
#> [1] "104.0473"
#>
#> $super_class
#> [1] "Fatty Acyls"
#>
#> $main_class
#> [1] "Fatty acids"
#>
#> $sub_class
#> [1] "Hydroxy FA"
Created on 2023-10-17 with reprex v2.0.2
Thanks for your answer! But it still gives an error 
> library(jsonlite)
> item <- fromJSON('https://www.metabolomicsworkbench.org/rest/refmet/match/2-Hydroxybutyricacid/name/txt')
Error in open.connection(con, "rb") :
cannot open the connection to 'https://www.metabolomicsworkbench.org/rest/refmet/match/2-Hydroxybutyricacid/name/txt'
In addition: Warning message:
In open.connection(con, "rb") :
URL 'https://www.metabolomicsworkbench.org/rest/refmet/match/2-Hydroxybutyricacid/name/txt': status was 'SSL connect error'
> item
Error: object 'item' not found
I just navigated there by cutting and pasting the link as you quote above and got
Can you try the same? If still not working do you know how to use the terminal to ping or traceroute? There may be network congestion or some other blockage in between.
When I click on the link directly I get the same result as you, but the code in R demonstrates that
Error in utils::download.file(url, ref) :
cannot open URL 'https://www.metabolomicsworkbench.org/rest/refmet/match/2''-Deoxyguanosine5''-Monophosphate/name/txt'
I try to use the options function and changing the mirrors to solve it, but they don't work too.
options(download.file.method = "wininet")
So it really bothers me. It is too much for newbie 
Thank you very much for your answer. It gives me the motivation to solve this problem.
This is strange. If I do
library(jsonlite)
item <- fromJSON('https://www.metabolomicsworkbench.org/rest/refmet/match/2-Hydroxybutyricacid/name/txt')
item
#> $refmet_name
#> [1] "2-Hydroxybutyric acid"
#>
#> $formula
#> [1] "C4H8O3"
#>
#> $exactmass
#> [1] "104.0473"
#>
#> $super_class
#> [1] "Fatty Acyls"
#>
#> $main_class
#> [1] "Fatty acids"
#>
#> $sub_class
#> [1] "Hydroxy FA"
Can you give us your complete code and perhaps the output of
sessionInfo()
Thanks
Beats me. This is working for me.
library(jsonlite)
download.file("https://www.metabolomicsworkbench.org/rest/refmet/match/2-Hydroxybutyricacid/name/txt", "target.txt")
I think you should open an issue on GitHub with the package authors.
Thank you for your patient reply. I think there may be a problem with the configuration of my computer system. I have decided to replace my laptop, hoping that similar situation will not happen again.
Wish you a nice day.
1 Like
Sometimes that's the only way. To avoid new issues, I suggest that before installing anything else, do R and RStudio taking all of the default options. Test. Then re-create the rest of your working environment testing along the way. Good luck!