Hello everyone, looking for information on the internet about how to create a matrix correlation in R, I realized that in all the examples they compare the same variable to each other. In my case I seek to compare between two different databases, but that are related to each other.
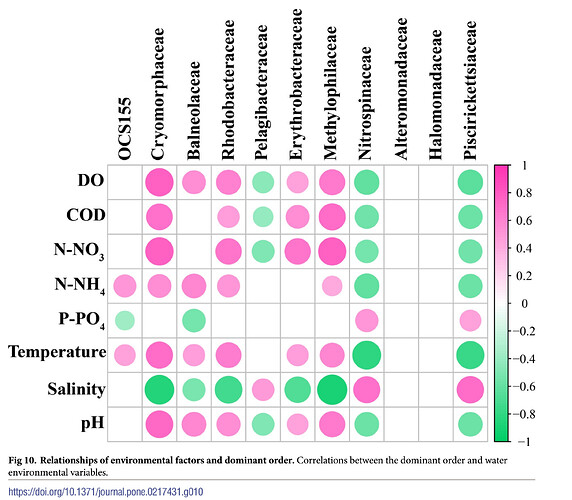
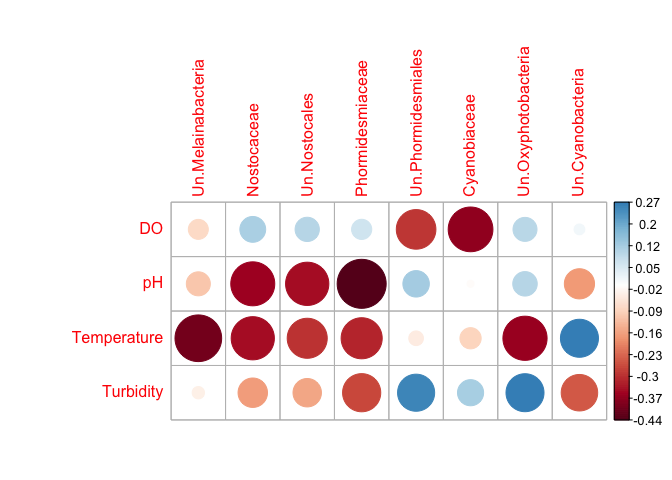
I found an example in a article of what type of correlation matrix i want to get:
My databases are very similar to the example above.
metadata <- data.frame(tibble::tribble(
~SampleID, ~DO, ~pH, ~Temperature, ~Turbidity,
"1A", 4.24, 9.94, 24.48, 87.3,
"1B", 2.58, 10.06, 24.95, 95.2,
"1C", 2.85, 9.98, 24.01, 98.8,
"1D", 3.81, 9.07, 26.67, 24.9,
"1E", 2.64, 8.99, 24.01, 43.1,
"1F", 4.61, 8.93, 23.5, 49.1,
"1G", 5.74, 9.24, 24.46, 68.6,
"1H", 3.69, 9.32, 24.65, 69.8,
"2A", 4.24, 9.94, 24.48, 87.3,
"2B", 2.66, 10, 24.92, 113,
"2C", 2.85, 9.98, 24.01, 98.8,
"2D", 3.81, 9.07, 26.67, 24.9,
"2E", 2.64, 8.99, 24.01, 43.1,
"2F", 4.61, 8.93, 23.5, 49.1,
"2G", 5.74, 9.24, 24.46, 68.6,
"2H", 3.69, 9.32, 24.65, 69.8
)
)
bacteria <- data.frame (tibble::tribble(
~index, ~Un.Melainabacteria, ~Nostocaceae, ~Un.Nostocales, ~Phormidesmiaceae, ~Un.Phormidesmiales, ~Cyanobiaceae, ~Un.Oxyphotobacteria, ~Un.Cyanobacteria,
"1A", 0, 0, 12, 66, 0, 0, 7137, 1467,
"1B", 0, 0, 6, 37, 0, 0, 5549, 238,
"1C", 0, 19, 0, 96, 0, 0, 1861, 708,
"1D", 0, 0, 0, 66, 0, 19, 3185, 4001,
"1E", 273, 1662, 207, 720, 67, 529, 7468, 1928,
"1F", 470, 2090, 425, 563, 30, 23, 18421, 4460,
"1G", 0, 372, 0, 19, 0, 2, 4191, 361,
"1H", 0, 433, 46, 1261, 33, 0, 12240, 1970,
"2A", 363, 1000, 8, 195, 37, 2, 20728, 4350,
"2B", 110, 769, 62, 46, 108, 648, 13438, 3076,
"2C", 169, 639, 31, 14, 11, 21, 17664, 2008,
"2D", 0, 510, 0, 118, 14, 0, 5085, 2745,
"2E", 129, 319, 0, 60, 0, 200, 8376, 1613,
"2F", 0, 501, 89, 818, 0, 0, 8906, 983,
"2G", 9, 930, 115, 167, 16, 195, 13273, 1514,
"2H", 209, 1876, 535, 316, 14, 43, 18506, 3531
)
)
Created on 2019-10-31 by the reprex package (v0.3.0)