Hi everyone,
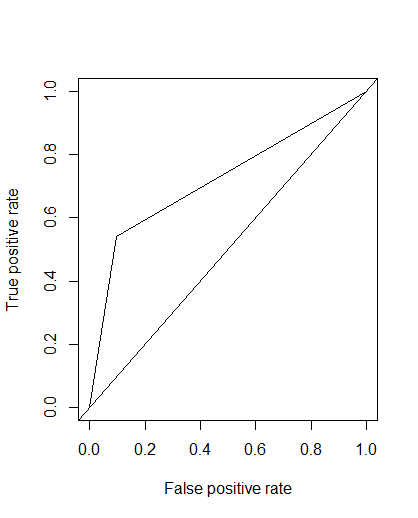
I am confused with a concept of getting the best threshold value for sensitivity and specificity.Below is my ROC curve.
It is obvious that my model is not good decision maker and this is because i have got very low sensitivity only (53%), Although i have got 0.80 aaccuracy.If i calculate threshold, it shows me some unethical cut-off.I will share my code below. From my knowledge, roc curve is the ratio between tpr and fpr and this tpr and fpr are depend on the threshold value.If i change the threshold value, these tpr and fpr should changed (Please rectify if i am wrong).So, my question is how can i calculate the best threshold value which keep the consistency between tpr and fpr. I know default cut-off is 0.5.But it can be varied.
I am going to reprex my full code so that you can understand,
library(ggplot2)
library(MASS)
library(car)
#> Loading required package: carData
library(mlogit)
#> Loading required package: Formula
#> Loading required package: maxLik
#> Loading required package: miscTools
#>
#> Please cite the 'maxLik' package as:
#> Henningsen, Arne and Toomet, Ott (2011). maxLik: A package for maximum likelihood estimation in R. Computational Statistics 26(3), 443-458. DOI 10.1007/s00180-010-0217-1.
#>
#> If you have questions, suggestions, or comments regarding the 'maxLik' package, please use a forum or 'tracker' at maxLik's R-Forge site:
#> https://r-forge.r-project.org/projects/maxlik/
library(sqldf)
#> Warning: package 'sqldf' was built under R version 3.5.1
#> Loading required package: gsubfn
#> Warning: package 'gsubfn' was built under R version 3.5.1
#> Loading required package: proto
#> Warning: package 'proto' was built under R version 3.5.1
#> Loading required package: RSQLite
#> Warning: package 'RSQLite' was built under R version 3.5.1
library(Hmisc)
#> Warning: package 'Hmisc' was built under R version 3.5.1
#> Loading required package: lattice
#> Loading required package: survival
#>
#> Attaching package: 'Hmisc'
#> The following objects are masked from 'package:base':
#>
#> format.pval, units
setwd("C:\\Users\\USER 1\\Downloads\\R File")
mydatacv <- read.csv("Telco-Customer-Churn.csv",header = TRUE)
str(mydatacv)
#> 'data.frame': 7043 obs. of 20 variables:
#> $ gender : Factor w/ 2 levels "Female","Male": 1 2 2 2 1 1 2 1 1 2 ...
#> $ SeniorCitizen : int 0 0 0 0 0 0 0 0 0 0 ...
#> $ Partner : Factor w/ 2 levels "No","Yes": 2 1 1 1 1 1 1 1 2 1 ...
#> $ Dependents : Factor w/ 2 levels "No","Yes": 1 1 1 1 1 1 2 1 1 2 ...
#> $ tenure : int 1 34 2 45 2 8 22 10 28 62 ...
#> $ PhoneService : Factor w/ 2 levels "No","Yes": 1 2 2 1 2 2 2 1 2 2 ...
#> $ MultipleLines : Factor w/ 3 levels "No","No phone service",..: 2 1 1 2 1 3 3 2 3 1 ...
#> $ InternetService : Factor w/ 3 levels "DSL","Fiber optic",..: 1 1 1 1 2 2 2 1 2 1 ...
#> $ OnlineSecurity : Factor w/ 3 levels "No","No internet service",..: 1 3 3 3 1 1 1 3 1 3 ...
#> $ OnlineBackup : Factor w/ 3 levels "No","No internet service",..: 3 1 3 1 1 1 3 1 1 3 ...
#> $ DeviceProtection: Factor w/ 3 levels "No","No internet service",..: 1 3 1 3 1 3 1 1 3 1 ...
#> $ TechSupport : Factor w/ 3 levels "No","No internet service",..: 1 1 1 3 1 1 1 1 3 1 ...
#> $ StreamingTV : Factor w/ 3 levels "No","No internet service",..: 1 1 1 1 1 3 3 1 3 1 ...
#> $ StreamingMovies : Factor w/ 3 levels "No","No internet service",..: 1 1 1 1 1 3 1 1 3 1 ...
#> $ Contract : Factor w/ 3 levels "Month-to-month",..: 1 2 1 2 1 1 1 1 1 2 ...
#> $ PaperlessBilling: Factor w/ 2 levels "No","Yes": 2 1 2 1 2 2 2 1 2 1 ...
#> $ PaymentMethod : Factor w/ 4 levels "Bank transfer (automatic)",..: 3 4 4 1 3 3 2 4 3 1 ...
#> $ MonthlyCharges : num 29.9 57 53.9 42.3 70.7 ...
#> $ TotalCharges : num 29.9 1889.5 108.2 1840.8 151.7 ...
#> $ Churn : int 0 0 1 0 1 1 0 0 1 0 ...
mydatacv$SeniorCitizen <- as.factor(mydatacv$SeniorCitizen)
mydatacv$Churn <- as.factor(mydatacv$Churn)
## Check Missing Value
sapply(mydatacv,function(x)sum(is.na(x)))
#> gender SeniorCitizen Partner Dependents
#> 0 0 0 0
#> tenure PhoneService MultipleLines InternetService
#> 0 0 0 0
#> OnlineSecurity OnlineBackup DeviceProtection TechSupport
#> 0 0 0 0
#> StreamingTV StreamingMovies Contract PaperlessBilling
#> 0 0 0 0
#> PaymentMethod MonthlyCharges TotalCharges Churn
#> 0 0 11 0
## Imputing missing value by mean through Hmisc package
library(Hmisc)
mydatacv$TotalCharges <- with(mydatacv,impute(TotalCharges,mean))
## Missing value imputation by mean without package
##mydata$TotalCharges <- ifelse(is.na(mydata$TotalCharges),
## mean(mydata$TotalCharges, na.rm=TRUE), mydata$TotalCharges)
## Check outlier
boxplot(mydatacv$tenure)
boxplot(mydatacv$MonthlyCharges)
boxplot(as.numeric(mydatacv$TotalCharges))
## Partition the data into two parts,(80%) and (20%)
set.seed(1234)
ind <- sample(2,nrow(mydatacv),replace=T,prob = c(0.8,0.2))
traindata <- mydatacv[ind==1,]
testdata <- mydatacv[ind==2,]
## K- fold Cross Validation
library(caret)
#>
#> Attaching package: 'caret'
#> The following object is masked from 'package:survival':
#>
#> cluster
controlparameter <- trainControl(method="cv",number=10,savePredictions = TRUE
)
##Apply Logistic Regression with K fold Cross Validation
##?train
modelglm <- train(Churn~.,data = traindata,method="glm",
family="binomial",trControl=controlparameter)
#> Warning in predict.lm(object, newdata, se.fit, scale = 1, type =
#> ifelse(type == : prediction from a rank-deficient fit may be misleading
#> Warning in predict.lm(object, newdata, se.fit, scale = 1, type =
#> ifelse(type == : prediction from a rank-deficient fit may be misleading
#> Warning in predict.lm(object, newdata, se.fit, scale = 1, type =
#> ifelse(type == : prediction from a rank-deficient fit may be misleading
#> Warning in predict.lm(object, newdata, se.fit, scale = 1, type =
#> ifelse(type == : prediction from a rank-deficient fit may be misleading
#> Warning in predict.lm(object, newdata, se.fit, scale = 1, type =
#> ifelse(type == : prediction from a rank-deficient fit may be misleading
#> Warning in predict.lm(object, newdata, se.fit, scale = 1, type =
#> ifelse(type == : prediction from a rank-deficient fit may be misleading
#> Warning in predict.lm(object, newdata, se.fit, scale = 1, type =
#> ifelse(type == : prediction from a rank-deficient fit may be misleading
#> Warning in predict.lm(object, newdata, se.fit, scale = 1, type =
#> ifelse(type == : prediction from a rank-deficient fit may be misleading
#> Warning in predict.lm(object, newdata, se.fit, scale = 1, type =
#> ifelse(type == : prediction from a rank-deficient fit may be misleading
#> Warning in predict.lm(object, newdata, se.fit, scale = 1, type =
#> ifelse(type == : prediction from a rank-deficient fit may be misleading
modelglm
#> Generalized Linear Model
#>
#> 5630 samples
#> 19 predictor
#> 2 classes: '0', '1'
#>
#> No pre-processing
#> Resampling: Cross-Validated (10 fold)
#> Summary of sample sizes: 5068, 5066, 5067, 5067, 5067, 5068, ...
#> Resampling results:
#>
#> Accuracy Kappa
#> 0.8023008 0.4614616
## Make prediction with mymodelglm to train data
prediction <- predict(modelglm,traindata)
#> Warning in predict.lm(object, newdata, se.fit, scale = 1, type =
#> ifelse(type == : prediction from a rank-deficient fit may be misleading
traindata$prediction <- prediction
##Validation of Prediction
confu_mat <- table(prediction=prediction,acctual=traindata$Churn)
confu_mat
#> acctual
#> prediction 0 1
#> 0 3732 687
#> 1 405 806
sensitivity <- 806/(806+687)
specificity <- 3732/ (3732+405)
print(c(specificity,sensitivity))
#> [1] 0.9021030 0.5398526
library(ROCR)
#> Loading required package: gplots
#>
#> Attaching package: 'gplots'
#> The following object is masked from 'package:stats':
#>
#> lowess
realvec <- ifelse(traindata$Churn==1,1,0)
predvec <- ifelse(prediction==1,1,0)
pred <- prediction(predvec,realvec)
roc1 <- performance(pred,"tpr","fpr")
plot(roc1)
abline(a=0,b=1)
##calculating threshold
eval=performance(pred,"acc")
plot(eval)
max_value<- which.max(slot(eval,"y.values")[[1]])
acc <- slot(eval,"y.values")[[1]][max_value]
thres <- slot(eval,"x.values")[[1]][max_value]
print(c(accuacy=acc,threshold_or_cutoff=thres))
#> accuacy threshold_or_cutoff
#> 0.8060391 1.0000000
## Calculaing AUC
auc <- performance(pred,"auc")
auc <- unlist(slot(auc,"y.values"))
auc <- round(auc,4)
auc
#> [1] 0.721
##Calculating miss-classification error
1-sum(diag(confu_mat))/sum(confu_mat) ##Accuracy
#> [1] 0.1939609
## Apply model on Test Data
pred_test <- predict(modelglm,newdata=testdata)
#> Warning in predict.lm(object, newdata, se.fit, scale = 1, type =
#> ifelse(type == : prediction from a rank-deficient fit may be misleading
testdata$prediction <- pred_test
confu_mat_test <- table(prediction=pred_test,actual=testdata$Churn)
confu_mat_test
#> actual
#> prediction 0 1
#> 0 920 157
#> 1 117 219
accuracy <- sum(diag(confu_mat_test))/sum(confu_mat_test)
misclass_test <- 1-accuracy
I am using K fold cross validation for developing the logistic regression model.
Another question i have,
when we develop any cross-validation , should we remove those variable whose p-value is less than level of significance or it automatically deals with those variables?
Any suggestion is really appreciable.
Thanks,
snandy