Hi everybody how are you?.
I would be very grateful if you could help me solve this.
I need to calculate Chao1 indixe for each sample and combines with the metadata, specifically with the months.
I already calculate other diversity indeces, and everything was fine, until I try to calculate Chao1.
My script for an example index: SIMPSON:
library(vegan)
library(readxl)
#rarefy con la media de reads por muestra Evaluation of Subsampling-Based Normalization Strategies for Tagged High-Throughput Sequencing Data Sets from Gut Microbiomes - PMC
genero<- read_excel("~/RSTUDIO/Datos_cianobacterias/Cianobacterias_genero_sin3h5c.xlsx")
#columna con las muestrasias
data <- genero
replicates <- as.data.frame(colnames(data)[-1])
colnames(replicates) <- "replicates"
attach(genero)
rwnames <- index
data <- as.matrix(data[,-1])
rownames(data) <- rwnames
data[is.na(data)] <- 0
rarefy(data, 94)
totales <- rowSums(data)
totales <- unname(totales)
mediana <- rowSums(data)
mediana <- unname(totales)
submuestras10 <- rrarefy(data, mediana)
write.csv(submuestras10, file = "~/RSTUDIO/Datos_cianobacterias/Cianobacterias_genero_rarefired_sin3h5c.csv")
#muestras en una fila
genero <- read.csv("~/RSTUDIO/Datos_cianobacterias/Cianobacterias_genero_rarefired_sin3h5c.csv", row.names=1)
data <- submuestras10
#data <- data[,colMeans(data) >=.1]
tot <- rowSums(data) #30 están no rarefied
simpson <- diversity(data, index = "simpson", base = exp(1))
write.csv(simpson, file = "~/RSTUDIO/Datos_cianobacterias/Cianobacterias_genero_rarefired_simpson_sin3h5c.csv")
#boxplot
#agregar el Tx= Control MS o T2DM
bpdata <- read.csv("~/RSTUDIO/Datos_cianobacterias/Metadata-final-sin3h5c.csv", row.names=1)
attach(bpdata)
colvec<- c("yellowgreen","darkturquoise", "coral")
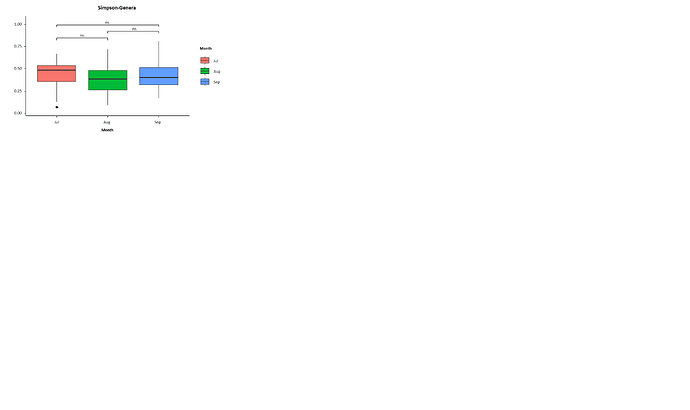
boxplot(simpson~Month, bpdata, col = colvec, ylab = "Simpson", main = "Species Diversity")
library(ggplot2)
library(ggsignif)
res.aov =aov(simpson ~ Month, data = bpdata)
summary(res.aov)#p-value<0.05
TukeyHSD(res.aov)
bpdata$Month <- factor(bpdata$Month,
labels = c("Jul", "Aug", "Sep"))
tumeans <- aggregate(simpson ~ Month, bpdata, mean)
library(ggplot2)
library(ggsignif)
library(tidyverse)
bxplot <- ggplot(bpdata, aes(x=Month, y = simpson, fill =Month )) +
geom_boxplot() +
geom_signif(comparisons = list(c("Jul", "Aug")),
map_signif_level=TRUE,
margin_top = 0.05)+
geom_signif(comparisons = list(c("Aug", "Sep")),
map_signif_level=TRUE,
margin_top = 0.15)+
geom_signif(comparisons = list(c("Jul", "Sep")),
map_signif_level=TRUE,
margin_top = 0.25)+
geom_boxplot(show.legend = FALSE) +
scale_y_continuous(expand = c(0.05,0.05))+
ggtitle("Simpson")+
theme_bw() +
theme(panel.grid.major = element_line(colour = "white"),
panel.grid.minor = element_blank(),
panel.border = element_blank(),
panel.background = element_blank(),
plot.title = element_text(hjust = 0.5, size = 14, family = "Tahoma", face = "bold"),
text=element_text(family = "Tahoma"),
axis.title = element_text(face="bold"),
axis.text.x = element_text(colour="black", size = 11),
axis.text.y = element_text(colour="black", size = 9),
axis.line = element_line(size=0.5, colour = "black"))
scale_fill_brewer(palette = "Accent")
bxplot
My reproducible data:
metadata <- data.frame(data.frame(stringsAsFactors=FALSE,
SampleID = c("1A", "1B", "1C", "1D", "1E", "1F", "1G", "1H", "2A",
"2B", "2C", "2D", "2E", "2F",
"2G", "2H", "3A", "3B", "3C", "3D",
"3E", "3F", "3G", "4A", "4B", "4C",
"4D", "4E", "4F", "4G", "4H", "5A",
"5B", "5D", "5E", "5F", "5G", "5H",
"6A", "6B", "6C", "6D", "6E", "6F",
"6G", "6H", "7A", "7B", "7C", "7D",
"7E", "7F", "7G", "7H", "8A", "8B",
"8C", "8D", "8E", "8F", "8G", "8H",
"9A", "9B", "9C", "9D", "9E", "9F",
"9G", "9H", "10A", "10B", "10C", "10D",
"10E", "10F", "10G", "10H", "11A",
"11B", "11C", "11D", "11E", "11F",
"11G", "11H", "12A", "12B", "12C",
"12D", "12E", "12F", "12G", "12H"),
SamplingPoint = c("CAJ-1", "CAJ-2", "CAJ-4", "CAJ-1", "CAJ-3", "CAJ-5",
"CAJ-2", "CAJ-4", "CAJ-1",
"CAJ-2", "CAJ-4", "CAJ-1", "CAJ-3",
"CAJ-5", "CAJ-2", "CAJ-4", "CAJ-1",
"CAJ-2", "CAJ-4", "CAJ-1", "CAJ-3",
"CAJ-5", "CAJ-2", "CAJ-1", "CAJ-3",
"CAJ-4", "CAJ-1", "CAJ-3", "CAJ-5",
"CAJ-2", "CAJ-4", "CAJ-1", "CAJ-3",
"CAJ-1", "CAJ-3", "CAJ-5", "CAJ-2",
"CAJ-4", "CAJ-1", "CAJ-3", "CAJ-5",
"CAJ-1", "CAJ-3", "CAJ-5", "CAJ-2",
"CAJ-4", "CAJ-1", "CAJ-3", "CAJ-5",
"CAJ-2", "CAJ-4", "CAJ-1", "CAJ-3",
"CAJ-5", "CAJ-1", "CAJ-3", "CAJ-5",
"CAJ-2", "CAJ-4", "CAJ-1", "CAJ-3",
"CAJ-5", "CAJ-2", "CAJ-3", "CAJ-5",
"CAJ-2", "CAJ-4", "CAJ-1", "CAJ-3",
"CAJ-5", "CAJ-2", "CAJ-3", "CAJ-5",
"CAJ-2", "CAJ-4", "CAJ-1", "CAJ-3",
"CAJ-5", "CAJ-2", "CAJ-4", "CAJ-5",
"CAJ-2", "CAJ-4", "CAJ-1", "CAJ-3",
"CAJ-5", "CAJ-2", "CAJ-4", "CAJ-1",
"CAJ-2", "CAJ-4", "CAJ-1", "CAJ-3",
"CAJ-5"),
Depth = c("80 cm", "80 cm", "Interstitial", "80 cm", "80 cm",
"80 cm", "80 cm", "80 cm",
"80 cm", "Interstitial", "Interstitial",
"80 cm", "80 cm", "80 cm", "80 cm",
"80 cm", "80 cm", "Interstitial",
"80 cm", "80 cm", "80 cm", "80 cm",
"80 cm", "Interstitial", "80 cm",
"80 cm", "80 cm", "80 cm", "80 cm",
"Interstitial", "Interstitial", "80 cm",
"80 cm", "80 cm", "80 cm", "80 cm",
"80 cm", "80 cm", "80 cm", "80 cm",
"80 cm", "80 cm", "80 cm", "80 cm",
"80 cm", "Interstitial",
"Interstitial", "Interstitial", "80 cm",
"80 cm", "80 cm", "80 cm", "80 cm",
"80 cm", "80 cm", "80 cm", "80 cm",
"80 cm", "80 cm", "80 cm", "80 cm",
"80 cm", "80 cm", "80 cm", "Interstitial",
"Interstitial", "Interstitial",
"Interstitial", "Interstitial",
"Interstitial", "80 cm", "80 cm", "80 cm",
"80 cm", "80 cm", "80 cm", "80 cm",
"Interstitial", "Interstitial",
"80 cm", "Interstitial", "80 cm",
"80 cm", "80 cm", "80 cm", "80 cm",
"Interstitial", "80 cm", "Interstitial",
"Interstitial", "Interstitial",
"Interstitial", "Interstitial",
"Interstitial"),
Month = c("July", "July", "July", "August", "August", "August",
"September", "September", "July",
"July", "July", "August", "August",
"August", "September", "September",
"July", "July", "July", "August",
"August", "August", "September",
"July", "July", "July", "August",
"August", "August", "September",
"September", "July", "July", "August",
"August", "August", "September",
"September", "July", "July", "July", "August",
"August", "August", "September",
"September", "July", "July", "July",
"August", "August", "September",
"September", "September", "July",
"July", "July", "August", "August",
"September", "September", "September",
"July", "July", "July", "August",
"August", "September", "September",
"September", "July", "July", "July",
"August", "August", "September",
"September", "September", "July", "July",
"July", "August", "August",
"September", "September", "September",
"July", "July", "August", "August",
"August", "September", "September",
"September")
)
)
results <- data.frame(data.frame(stringsAsFactors=FALSE,
index = c("1A", "1B", "1C", "1D", "1E", "1F", "1G", "1H",
"2A", "2B", "2C", "2D",
"2E", "2F", "2G", "2H", "3A",
"3B", "3C", "3D", "3E",
"3F", "3G", "4A", "4B", "4C",
"4D", "4E", "4F", "4G", "4H",
"5A", "5B", "5D", "5E",
"5F", "5G", "5H", "6A", "6B",
"6C", "6D", "6E", "6F", "6G",
"6H", "7A", "7B", "7C",
"7D", "7E", "7F", "7G", "7H",
"8A", "8B", "8C", "8D", "8E",
"8F", "8G", "8H", "9A",
"9B", "9C", "9D", "9E", "9F",
"9G", "9H", "10A", "10B",
"10C", "10D", "10E", "10F",
"10G", "10H", "11A", "11B",
"11C", "11D", "11E", "11F",
"11G", "11H", "12A", "12B",
"12C", "12D", "12E", "12F",
"12G", "12H"),
Un.Caenarcaniphilales = c(0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0,
0, 0, 0, 0, 0, 0, 0, 0, 0,
0, 0, 0, 0, 0, 0, 0, 0, 0,
0, 0, 0, 0, 0, 0, 0, 0, 0,
0, 0, 0, 0, 0, 0, 0, 0, 0, 0,
0, 0, 0, 0, 0, 0, 0, 2, 0,
0, 0, 0, 0, 0, 0, 0, 0, 0,
0, 0, 0, 0, 0, 0, 0, 0, 0, 0,
0, 0, 0, 0, 0, 0, 0, 0, 2,
0, 0, 0, 0),
Un.Gastranaerophilales = c(0, 2, 0, 0, 0, 27, 0, 0, 22, 0, 0, 41, 0, 0, 9,
11, 0, 0, 0, 0, 0, 0, 0,
203, 0, 0, 28, 0, 0, 2, 15,
0, 0, 0, 0, 8, 0, 0, 9, 0, 0,
0, 0, 31, 0, 0, 0, 0, 0, 0,
2, 0, 0, 0, 19, 8, 0, 24,
0, 12, 0, 0, 0, 0, 0, 8, 0,
0, 0, 0, 0, 0, 38, 0, 0, 0,
0, 0, 0, 0, 0, 14, 0, 0, 0,
0, 0, 0, 21, 0, 0, 40, 0, 3),
Un.Obscuribacterales = c(0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0,
0, 0, 0, 0, 0, 0, 0, 0, 0,
0, 0, 0, 0, 0, 0, 0, 0, 0,
0, 0, 0, 0, 0, 0, 0, 0, 0,
0, 0, 0, 0, 0, 0, 0, 0, 0, 0,
0, 0, 0, 0, 0, 0, 0, 0, 0,
0, 0, 0, 0, 0, 0, 0, 0, 0,
0, 0, 0, 0, 0, 0, 0, 0, 0, 0,
0, 0, 0, 0, 0, 0, 0, 2, 0,
0, 0, 0, 0),
Un.Vampirovibrionales = c(0, 0, 0, 300, 0, 0, 0, 0, 221, 0, 0, 0, 282, 0,
0, 7, 232, 0, 0, 95, 94,
0, 0, 626, 0, 0, 0, 0, 0,
171, 0, 1139, 0, 0, 0, 0, 57,
0, 311, 1003, 0, 0, 119, 7,
99, 0, 422, 0, 0, 0, 0, 0,
289, 0, 322, 114, 0, 2, 5, 0,
340, 4, 0, 0, 98, 0, 0, 0,
0, 9, 121, 152, 0, 0, 0, 105,
0, 0, 0, 0, 0, 0, 2, 20, 0,
0, 48, 0, 0, 5, 0, 0, 0,
15),
Un.Melainabacteria = c(0, 0, 0, 0, 273, 470, 0, 0, 363, 110, 169, 0,
129, 0, 9, 209, 0, 0, 0,
0, 42, 64, 94, 1050, 154, 0,
209, 377, 0, 3, 0, 88, 0,
143, 0, 85, 90, 21, 491, 329,
45, 0, 215, 384, 0, 154, 21,
0, 0, 42, 2, 17, 7, 0, 291,
299, 27, 352, 74, 354, 241,
2, 0, 0, 0, 749, 0, 116, 0,
0, 163, 73, 0, 0, 386, 132,
38, 0, 0, 0, 0, 488, 176,
0, 0, 0, 0, 71, 40, 273, 0,
534, 485, 0),
Limnotrichaceae = c(0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0,
0, 0, 0, 0, 0, 0, 0, 0, 0,
0, 0, 0, 0, 0, 0, 0, 0, 0,
0, 0, 0, 0, 0, 0, 0, 0, 0,
0, 0, 0, 0, 0, 0, 0, 0, 0, 0,
0, 0, 0, 0, 0, 0, 0, 0, 0,
0, 0, 0, 0, 0, 0, 0, 2, 0,
0, 0, 0, 0, 0, 0, 0, 0, 0, 0,
0, 0, 0, 0, 0, 0, 0, 0, 0,
0, 0, 0, 0),
Cyanobacteriaceae = c(0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 3, 0, 0, 0, 0,
0, 0, 0, 0, 0, 0, 0, 0, 0,
0, 0, 0, 0, 0, 0, 0, 0, 0,
0, 0, 0, 0, 0, 0, 0, 0, 0,
0, 0, 0, 0, 0, 0, 0, 0, 0, 0,
0, 0, 0, 0, 0, 0, 0, 0, 0,
0, 0, 0, 0, 0, 0, 0, 0, 0,
0, 0, 0, 0, 0, 0, 0, 0, 0, 0,
0, 0, 0, 0, 0, 0, 0, 0, 0,
0, 0, 0, 0),
Microcystaceae = c(0, 0, 0, 0, 0, 2, 0, 112, 0, 0, 0, 0, 0, 3, 0, 0,
0, 0, 0, 0, 0, 2, 0, 7, 0,
0, 0, 3, 0, 0, 3, 0, 0, 0,
0, 5, 2, 0, 0, 0, 0, 0, 0,
0, 0, 55, 0, 0, 0, 0, 0, 2,
0, 0, 0, 0, 0, 3, 0, 0, 93,
35, 0, 0, 0, 0, 0, 2, 13,
396, 0, 0, 0, 0, 0, 0, 0, 0, 0,
0, 0, 0, 2, 0, 0, 0, 7, 0,
0, 20, 0, 0, 70, 222),
Nostocaceae = c(0, 0, 19, 0, 1662, 2090, 372, 433, 1000, 769,
639, 510, 319, 501, 930,
1876, 316, 613, 0, 0, 1039,
388, 982, 3272, 895, 243,
1621, 1094, 43, 291, 2830, 2,
0, 1006, 2, 1051, 1042, 244,
1258, 1190, 266, 63, 1016,
2226, 206, 2014, 183, 0, 27,
139, 1266, 232, 1036, 102,
1096, 2439, 39, 2214, 1709,
2633, 2602, 1403, 2, 5, 46,
3157, 342, 1031, 407, 5477,
654, 343, 358, 38, 2265, 824,
338, 151, 11, 27, 0, 1108,
1105, 0, 0, 88, 441, 404,
623, 3218, 172, 2631, 3524,
1832)
)
)
Created on 2019-10-07 by the reprex package (v0.3.0)
This is an example that i found on internet, and i like it, but I could'nt do it
Thanks in advance