Hi guys
I need help drawing a corrplot.
lee <- tibble::tribble(
~NAME, ~`Bacteria-1`, ~`Bacteria-2`, ~`Bacteria-3`, ~`Bacteria-4`, ~`Bacteria-5`, ~`Bacteria-6`, ~`Bacteria-7`, ~`Bacteria-8`, ~`Bacteria-9`, ~`Bacteria-10`, ~`Bacteria-11`, ~`Bacteria-12`, ~`Bacteria-13`, ~`Bacteria-14`,
"CON-1", 41L, 13L, 3293L, 22L, 24L, 59L, 38L, 111L, 46L, 5L, 164L, 47L, 49L, 21L,
"CON-2", 46L, 11L, 3099L, 33L, 27L, 66L, 42L, 98L, 37L, 8L, 170L, 50L, 60L, 26L,
"CON-3", 58L, 12L, 3248L, 12L, 24L, 73L, 45L, 97L, 39L, 14L, 169L, 43L, 52L, 20L
)
type or paste code here
kwon <- tibble::tribble(
~NAME, ~pH, ~co2, ~ch4, ~o2, ~n2o, ~h2o,
"CON-1", 47.2, 6.29, 65.19, 60.09, 25.64, 14.27,
"CON-2", 48.2, 6.24, 67.76, 59.89, 26.77, 13.34,
"CON-3", 44.6, 6.28, 65.29, 59.75, 26.24, 14
)
Those are input data to drawing corrplot
and this is my code
library(corrplot)
library(tidyverse)
library(readxl)
library(Hmisc)
data1<-read.csv("raw_data/lee.csv",head=T)
data2<-read.csv("raw_data/kwon.csv", head=T)
cor_result<-matrix(NA, 6, 14)
pval_result<-matrix(NA, 6, 14)
%>%
for(i in 2:ncol(data1)){
for(j in 2:ncol(data2)){
output<- cor.test(data1[,i],data2[,j], method = "pearson", use="na.or.complete")
cor_result[j-1,i-1] <- output$estimate
pval_result[j-1,i-1] <- output$p.value
}}
colnames(cor_result) <- names(data1)[2:15]
rownames(cor_result) <- names(data2)[2:7]
colnames(pval_result) <- names(data1)[2:15]
rownames(pval_result) <- names(data2)[2:7]
cor_result[abs(cor_result) < 0.9] <- NA
pval_result[abs(cor_result) < 0.05] <-NA
corrplot(cor_result, method='circle',tl.cex=0.7, na.label=" ", tl.col="black", addCoef.col = 'white', number.cex = 1)
corrplot(pval_result, method='circle',tl.cex=0.7, na.label=" ", tl.col="black", addCoef.col = 'white', number.cex = 1)
I want to bind this pval_result and cor_result (together) to indicate corrplot as satisfied same times.
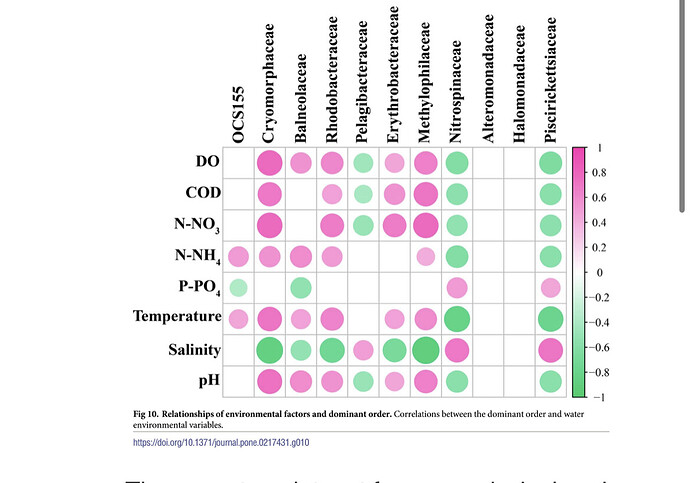
Some papers indicated that "pearson correlation coefficient (p<0.05, lrl>0.7)" were shown on the plot.
I also want to make plot like those conditions.
Please help me!