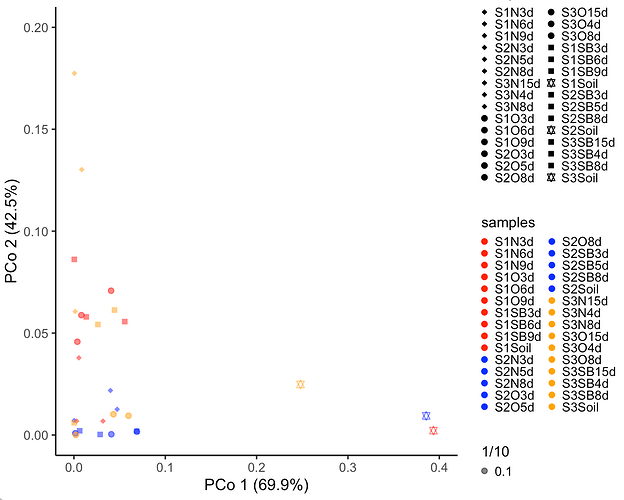
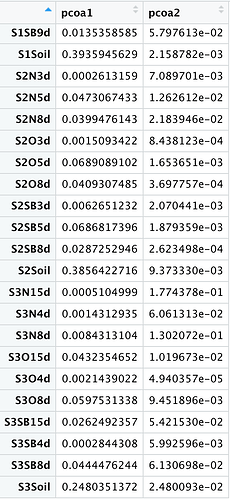
I created a scatter plot using the following script.
pcoa_plot <- positions %>%
as_tibble(rownames = "samples") %>%
ggplot(aes(x=pcoa1, y=pcoa2, shape= samples)) +
geom_point() +
aes(colour=samples) + #adds legend to the plot
labs(x="PCo 1 (69.9%)", y="PCo 2 (42.5%)") +
scale_color_manual(
values = c("S1N3d"= "red","S1N6d"= "red", "S1N9d"= "red", "S1O3d"= "red",
"S1O6d"= "red", "S1O9d"= "red", "S1SB3d"= "red", "S1SB6d"= "red",
"S1SB9d"= "red", "S1Soil" = "red",
"S2N3d"= "blue","S2N5d"= "blue", "S2N8d"= "blue", "S2O3d"= "blue",
"S2O5d"= "blue", "S2O8d"= "blue", "S2SB3d"= "blue", "S2SB5d"= "blue",
"S2SB8d"= "blue", "S2Soil" = "blue",
"S3N15d"= "orange","S3N4d"= "orange", "S3N8d"= "orange", "S3O15d"= "orange",
"S3O4d"= "orange", "S3O8d"= "orange", "S3SB15d"= "orange", "S3SB4d"= "orange",
"S3SB8d"= "orange", "S3Soil" = "orange")) +
scale_shape_manual(values = c("S1N3d"= "diamond", "S1N6d"= "diamond", "S1N9d"= "diamond",
"S2N3d"= "diamond","S2N5d"= "diamond", "S2N8d"= "diamond",
"S3N15d"= "diamond","S3N4d"= "diamond", "S3N8d"= "diamond",
"S1O3d"= "circle", "S1O6d"= "circle", "S1O9d"= "circle",
"S2O3d"= "circle", "S2O5d"= "circle", "S2O8d"= "circle",
"S3O15d"= "circle", "S3O4d"= "circle", "S3O8d"= "circle",
"S1SB3d"= "square", "S1SB6d"= "square", "S1SB9d"= "square", "S1Soil" = "star",
"S2SB3d"= "square", "S2SB5d"= "square", "S2SB8d"= "square", "S2Soil" = "star",
"S3SB15d"= "square", "S3SB4d"= "square", "S3SB8d"= "square", "S3Soil" = "star")) +
aes(alpha= 1/10) +
scale_x_continuous(limits = c(0.0, 0.4)) +
scale_y_continuous(limits = c(0.0, 0.2)) +
theme_void() +
theme_classic() +
theme(legend.key.size = unit(0.1, 'cm'), #change legend key size
legend.key.height = unit(0.001, 'cm'), #change legend key height
legend.key.width = unit(0.001, 'cm'),
legend.title = element_text(size=10))
I get the plot as follows
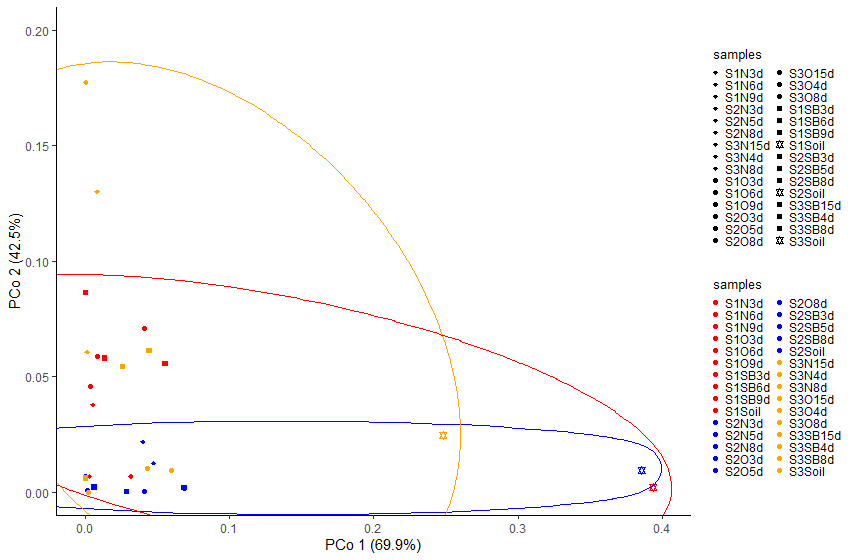
I want to create this scatter plot with ellipses in such a way that all red will be grouped in one ellipse, all blue in a separate ellipse and all yellow in separate ellipse.
Please help me in this regard. I hope i was able to explain my question well.
Regards
Hira