Hi community, I have this loop to extract only the filezise from many files. But the total files are like 300,000 with .hips format. So, in a simple rule of three I am getting all the time it is 5.15 days to run the code.
Is there any way to make this loop faster, maybe with map family functions?
Others?
With this function I first get all the directories, then I create the path entering the blobs folder and there I select only the .hips files. After that I capture only the size of each file.
library(tidyverse)
library(exiftoolr)
### TODOS DIRECTORIOS
rootPath = '//ALLIANCEDFS.ALLIANCE.CGIAR.ORG/CL07_GRPhistoric/GRP_OPERATION/SEEDS/Videometro/Blobs/BlobCollections Secundary & Tertiary color/'
list_dir_2 <- data.frame(root=list.dirs(rootPath)) |>
mutate(folder = gsub(rootPath, "", root))
list_dir_2_blobs <- list_dir_2 |>
filter(grepl("/blobs", folder)) |>
mutate(folder_final=paste0(root,'/'))
# function
get_hips_info <- function(directory) {
file_names <- list.files(path.expand(directory), pattern = "\\.hips$")
FileSize <- paste0(directory, file_names) %>%
lapply(function(x) {
cat("Reading file:", x, "\n") # Print a message when reading each file
exif_read(x)$FileSize
}) %>% unlist
hips <- data.frame(file = file_names, FileSize = FileSize) %>%
mutate(measure_1 = paste0('bytes'))
return(hips)
}
inicio <- Sys.time() # start
# fuction for each directory
result_list <- lapply(list_dir_2_blobs $folder_final, get_hips_info)
fin <- Sys.time() # fin
tiempo_ejecucion <- fin-inicio;tiempo_ejecucion # time of run
# dataframe
result_df <- bind_rows(result_list)
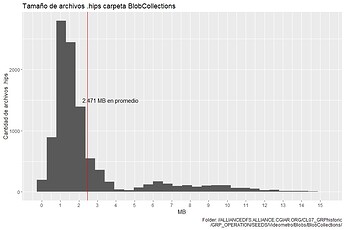
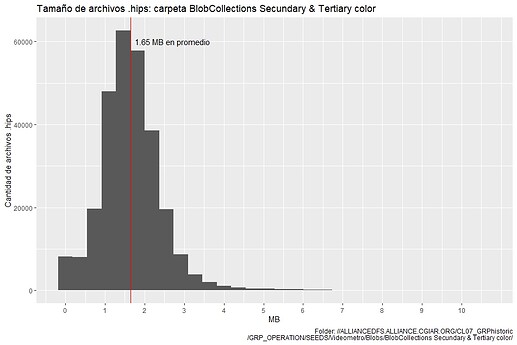
The histogram for the first 10265 .hips file:
Tnks!