First of all, your data fr the x-axis need to be numeric!
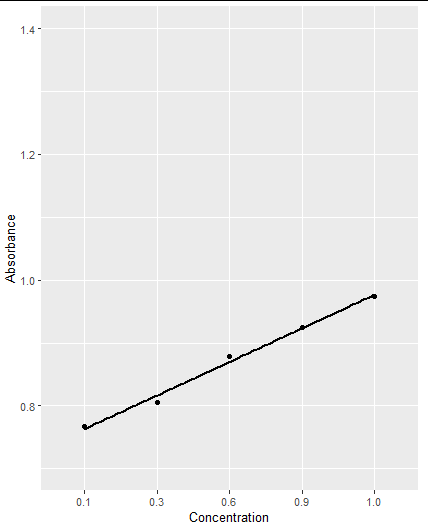
data2 = data.frame(Concentration = c(0.1, 0.3, 0.6, 0.9, 1.0),
Absorbance = c(0.767, 0.805, 0.879, 0.925, 0.974))
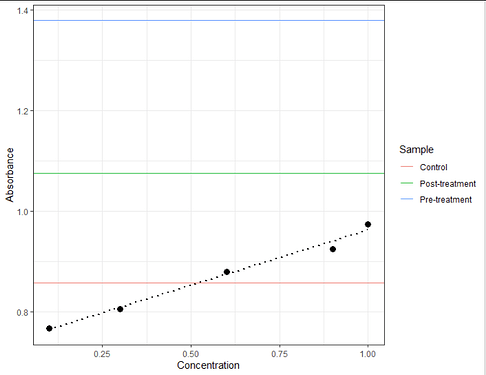
data3 = data.frame(Sample = c("Control", "Pre-treatment", "Post-treatment"),
Absorbance = c(0.857, 1.379, 1.075))
Then for the start you can just show the measured absorbances for the 3 samples as lines.
You see only control is within your calibration curve, post-treatment is a bit, pre-treatment massively outside.
This isn't good, as you don't know whether the trend remains linear or goes into a saturation.
#show the plot with readouts as line
ggplot(data = data2,
aes(x = Concentration, y = Absorbance)) +
geom_point(size = 3) +
stat_smooth(method=lm, se = FALSE,
colour = "black", linetype = "dotted") +
theme_bw() +
geom_hline(data = data3,
aes(yintercept = Absorbance,
colour = Sample))
Then you can calculate a fit trough your values:
#calculate the linear fit
linear_fit=lm(Absorbance ~ Concentration, data = data2)
summary(linear_fit)
#
#Call:
# lm(formula = Absorbance ~ Concentration, data = data2)
#
#Residuals:
# 1 2 3 4 5
#0.002388 -0.003524 0.004609 -0.015259 0.011786
#
#Coefficients:
# Estimate Std. Error t value Pr(>|t|)
#(Intercept) 0.74266 0.01029 72.20 5.86e-06 ***
# Concentration 0.21956 0.01527 14.38 0.000729 ***
# ---
# Signif. codes: 0 ‘***’ 0.001 ‘**’ 0.01 ‘*’ 0.05 ‘.’ 0.1 ‘ ’ 1
#
#Residual standard error: 0.01171 on 3 degrees of freedom
#Multiple R-squared: 0.9857, Adjusted R-squared: 0.9809
#F-statistic: 206.9 on 1 and 3 DF, p-value: 0.0007286
# good fit!
# extract fitted (= expected) y values
linear_fit$fitted.values
# 1 2 3 4 5
# 0.7646122 0.8085238 0.8743912 0.9402585 0.9622143
# this is quite close to what we put in
# extract the coeficicents
linear_fit$coefficients
# (Intercept) Concentration
# 0.7426565 0.2195578
I am sure there must be a function to calculate x from y, for now I just found the opposite, predict calculates y from x. But we can do the math on our own and write a function for this:
# write a function to define x from given y
find_x = function(model, y) {
# y = slope*x + intercept
# y - intercept = slope*x
# (y-intercept) / slope = x
# just extract both values and do the math:
intercept = model$coefficients[1]
slope = model$coefficients[2]
x = (y-intercept)/slope
return(x)
}
# check with inputs
find_x(linear_fit, linear_fit$fitted.values)
# 1 2 3 4 5
# 0.1 0.3 0.6 0.9 1.0
# okay works fine!
# calculate for data3
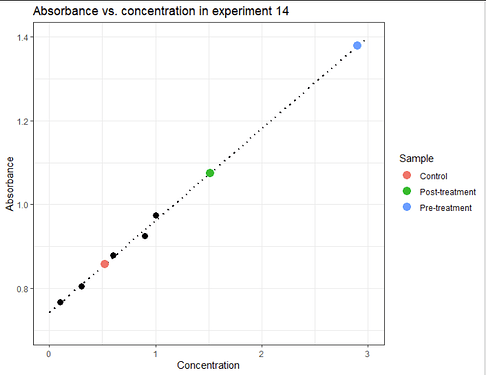
find_x(linear_fit, data3$Absorbance)
# [1] 0.5207901 2.8982959 1.5136948
Ultimately we can add the results to data_3 and show the fitted results on the plot:
# add result to data3
data3$Concentration = find_x(linear_fit, data3$Absorbance)
# plot again with points
ggplot(data = data2,
aes(x = Concentration, y = Absorbance)) +
geom_smooth(method=lm, se = FALSE,
colour = "black", linetype = "dotted",
fullrange = TRUE) + # to expnad it beyond the inputs
geom_point(size = 3) +
geom_point(data = data3,
aes(colour = Sample),
size = 4) +
ylim(0.7, 1.4) + xlim(0, 3) +
theme_bw() +
labs(title = "Absorbance vs. concentration in experiment 14") # or whatever title you need
![]()