Continuing the discussion from How can I set the right font of heatmap to be italic?And some font to be superscripts or subscript below heatmap?:
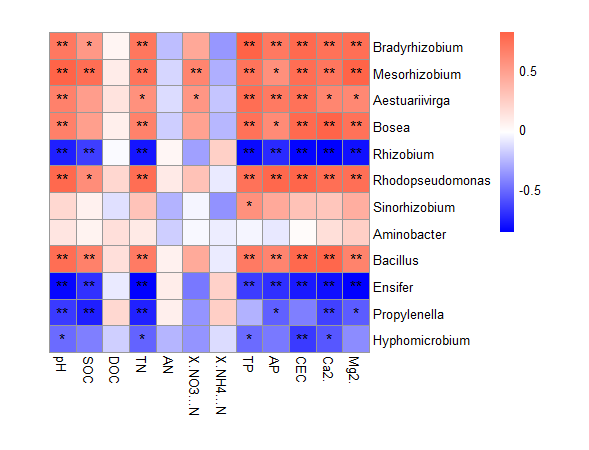
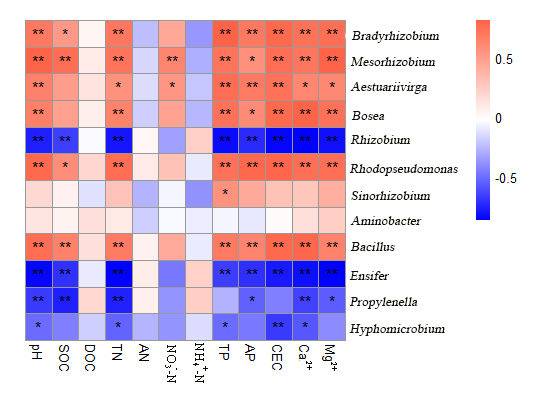
Thanks!But there are some problem.Fig.1 is prime.So how can I make the code?
(NO3-)-N is Nitrate Nitrogen and it should be the right font.3 should be subscript ,- after 3 should be superscript.
(NH4+)-N is Ammonium nitrogen and it should be the right font.4 should be subscript ,+ after 4 should be superscript.
Ga2+/Mg2+ The 2+ should be superscript.
The right font of heatmap should be italic.
setwd(choose.dir())
options(repos=structure(c(CRAN="The Comprehensive R Archive Network")))
install.packages('psych')
install.packages("reshape2")
library(psych)
library(corrplot)
library(pheatmap)
library(reshape2)
Order<-read.table('nirk_属水平.txt', header = TRUE,
sep = '\t',row.names = 1)
Order<-t(Order)
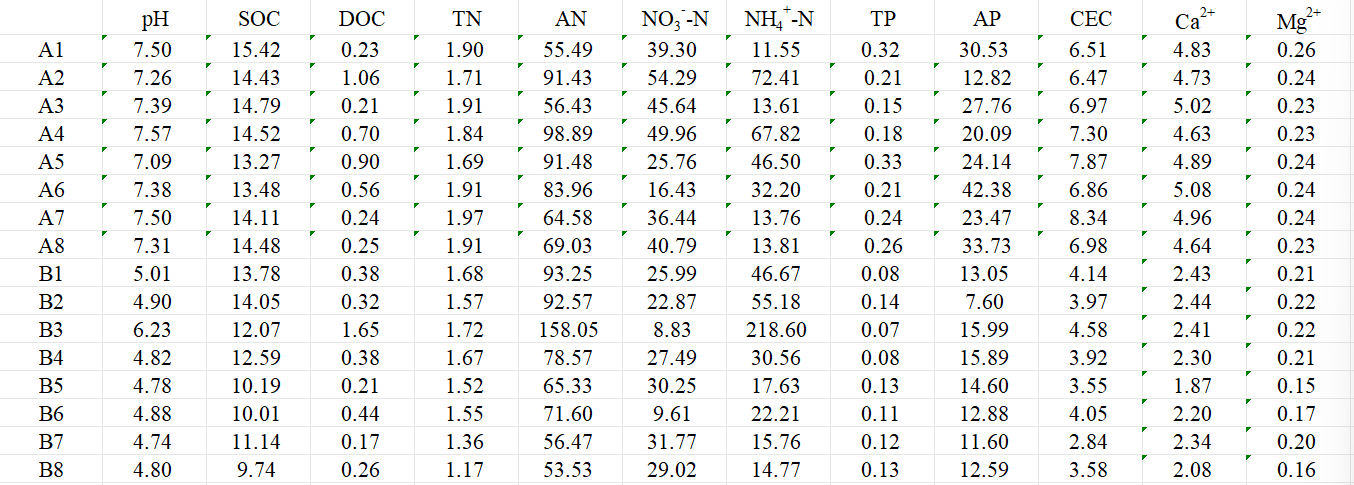
lihua<-read.table('env_table.txt', header = TRUE,
sep = '\t',row.names = 1)
res<-corr.test(Order, lihua, method = 'spearman',
adjust = 'holm',alpha = 0.05)
rmt<-res$r
pmt<-res$p
mycol<-colorRampPalette(c("blue","white","tomato"))(800)
if (!is.null(pmt)){ssmt <- pmt< 0.01
pmt[ssmt] <-'**'
smt <- pmt >0.01& pmt <0.05
pmt[smt] <- '*'
pmt[!ssmt&!smt]<- ''
} else {
pmt <- F
}
pheatmap(rmt, cluster_rows = F, cluster_cols = F,
display_numbers = pmt,fontsize_number = 12,
number_color = "black", cellwidth = 20,
cellheight = 20,color = mycol)