Here is where I'm at - starting with progress from your example:
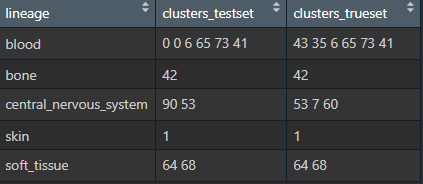
#Dataframe of lineage, testset, and truest
DF <- data.frame(lineage = c("blood", "bone", "central", "skin", "soft"),
clusters_testset = c("0 0 6 65 73 41", "42", "90 53", "1", "65 68"),
clusters_trueset = c("43 35 6 65 73 41", "42", "53 7 60", "73", "60 68"))
#str_split on both columns
library(stringr)
testset <- str_split(DF$clusters_testset, pattern = " ")
trueset <- str_split(DF$clusters_trueset, pattern = " ")
Ultimate goal: Make and attach to DF the following 4 columns:
(Using blood lineage as an example, but I need to do this for each row)
a_result = # of blood clusters in testset that DO match with blood clusters in trueset
b_result = # of blood clusters in testset that DO NOT match with blood clusters in trueset
c_result = # of any NON-blood clusters of testset that DO match with blood clusters in trueset
d_result = # any NON-blood clusters that DO NOT match with blood clusters in trueset (which will be the biggest number)
First manually find the correct answers to later check against:
DF
lineage clusters_testset clusters_trueset
1 blood 0 0 6 65 73 41 43 35 6 65 73 41
2 bone 42 42
3 central 90 53 53 7 60
4 skin 1 73
5 soft 65 68 60 68
Again, using just blood as the example:
a_result = 4 (4 blood in testset match with trueset)
b_result = 2 (2 testset blood clusters do no match anywhere with blood trueset)
c_result = 1 (Only 1 non-blood cluster in the testset column matches with one of the blood trueset clusters [it is skin, cluster 65])
d_result = 5 (There are 5 non-blood clusters in testset that do not match with blood clusters of trueset)
Approach so far:
a_result:
DF$a_result <- map2_dbl(testset, trueset, function(x, y) sum(x %in% y))
b_result:
According to this R-bloggers post you can define a %notin% operator to negate %in%:
`%notin%` <- Negate(`%in%`)
DF$b_result <- map2_dbl(testset, trueset, function(x, y) sum(x %notin% y))
So far, this works for a_result and b_result:
DF
lineage clusters_testset clusters_trueset a_result b_result
1 blood 0 0 6 65 73 41 43 35 6 65 73 41 4 2
2 bone 42 42 1 0
3 central 90 53 53 7 60 1 1
4 skin 1 73 0 1
5 soft 65 68 60 68 1 1
I can't seem to make anything work for c_result, d_result : any ideas?
Thanks for your help!