Hello everyone...
I am a master student stuck in a problem for half a day...
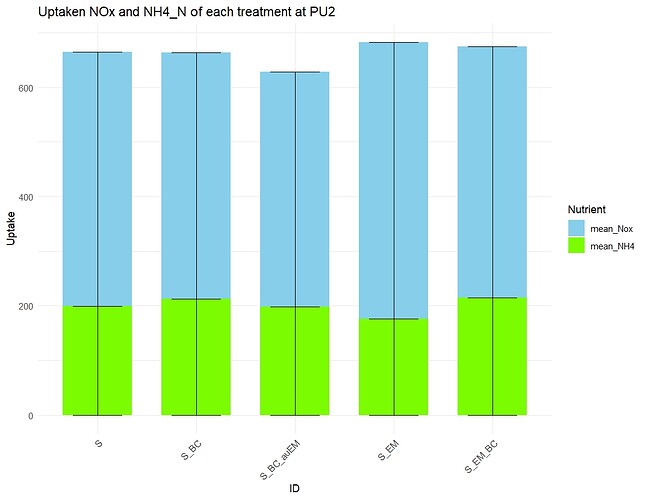
I am making a bar plot with the error bars.
I have 2 values which are NOx and NH4_N.
And there are 5 IDs which are S, S_BC, S_EM, S_BC_auEM, and S_EM_BC.
For the bar plot, I want to include NOx and NH4 data in one bar to indicate the Total N uptake. However, the inclusion of the bar worked quite well, but the error bar problem could not be solved.
like the are not stacked or looks so weird like here.
The code is the following...
####Trial
### Uptaken N - Pu2
# Calculate mean and standard error for Uptook_Nox and Uptook_NH4_N by ID
summary_stats_pu2 <- data_1_sub_tray_Pu2 %>%
group_by(ID) %>%
summarise(mean_Nox = mean(Uptook_Nox),
mean_NH4 = mean(Uptook_NH4_N),
se_Nox = sd(Uptook_Nox) / sqrt(n()),
se_NH4 = sd(Uptook_NH4_N) / sqrt(n()))
# Reshape data for plotting
plot_data_pu2 <- summary_stats_pu2 %>%
pivot_longer(cols = c(mean_Nox, mean_NH4), names_to = "Nutrient", values_to = "Mean") %>%
mutate(Error = ifelse(Nutrient == "mean_Nox", se_Nox, se_NH4),
Nutrient = factor(Nutrient, levels = c("mean_NH4", "mean_Nox"))) # Ensure NH4 is on top
# Plotting stacked bar chart with error bars
ggplot(plot_data_pu2, aes(x = ID, y = Mean, fill = Nutrient)) +
geom_bar(stat = "identity", position = position_stack(reverse = TRUE), width = 0.7) +
geom_errorbar(aes(ymin = Mean - Error, ymax = Mean + Error),
position = position_stack(reverse = TRUE, vjust = 0.5), width = 0.5, # Adjust position and width
size = 0.5, color = "black") + # Adjust error bar aesthetics
labs(x = "ID", y = "Uptake", fill = "Nutrient",
title = "Uptaken NOx and NH4_N of each treatment at PU2") +
theme_minimal() +
theme(axis.text.x = element_text(angle = 45, hjust = 1)) +
scale_fill_manual(values = c("mean_NH4" = "#7CFC00", "mean_Nox" = "skyblue")) + # Custom fill colors
guides(fill = guide_legend(reverse = TRUE)) # Reverse legend order to match stack order
What should I change to make the error bar proper?
Your idea will be highly appreciated... thank you in advance!