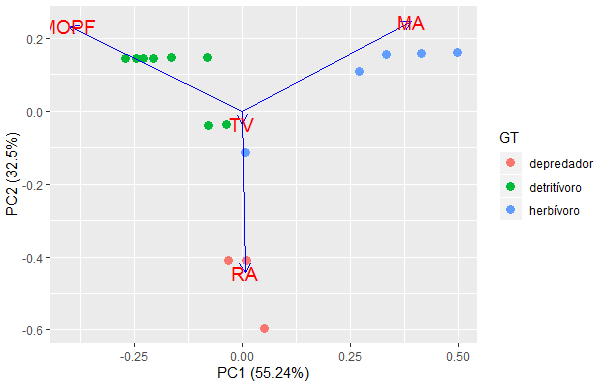
Hi, I made a PCA with ggplot2, however I want my PCA points to have a name, what function could I use. Thank you
Taxon MOPF MA RA TV GT
1 Andesiops 75 25 0 0 detritívoro
2 Meridialaris 5 95 0 0 herbívoro
3 Polycentropus 20 0 80 0 depredador
4 Austrelmis 93 7 0 0 detritívoro
5 Prionocyphon 40 0 60 0 depredador
6 Neoplasta 98 2 0 0 detritívoro
7 Gigantodax 65 15 20 0 detritívoro
8 Simulium 30 65 5 0 herbívoro
9 Tipula 30 0 0 70 herbívoro
10 Alotanypus 60 20 20 0 detritívoro
11 Pentaneura 35 5 60 0 depredador
12 Corynoneura 15 85 0 0 herbívoro
13 Cricotopus 90 10 0 0 detritívoro
14 Tanytarsus 95 5 0 0 detritívoro
15 Podonomus 25 75 0 0 herbívoro
16 Podonomopsis 85 15 0 0 detritívoro
17 Parochlus 95 5 0 0 detritívoro