Hi community
In this data I want to order acording with Valor. But each germplasmName have two times, SEMBRADAS and GERMINADAS.
library(tidyverse)
library(readxl)
library(forcats)
DF2 <-structure(list(germplasmName = c("AM206-5", "AM206-5", "ARG73",
"ARG73", "B1PD280-008", "B1PD280-008", "BRA847", "BRA847", "BRA97",
"BRA97", "C4", "C4", "CG1141-1", "CG1141-1", "CM4919-1", "CM4919-1",
"CM523-7", "CM523-7", "COL1468", "COL1468", "COL22", "COL22",
"COL2215", "COL2215", "CPDCR1B-052", "CPDCR1B-052", "CPDCR1B-080",
"CPDCR1B-080", "CPDCR5B-036", "CPDCR5B-036", "CPDCR5B-069A",
"CPDCR5B-069A", "CR63", "CR63", "GM3426-5", "GM3426-5", "GM3732-14",
"GM3732-14", "GM3732-17", "GM3732-17", "GM3732-21", "GM3732-21",
"GM3732-22", "GM3732-22", "GM3732-5", "GM3732-5", "GM3736-15",
"GM3736-15", "GM3736-16", "GM3736-16", "GM3736-20", "GM3736-20",
"GM3736-26", "GM3736-26", "GM3736-28", "GM3736-28", "GM3736-29",
"GM3736-29", "GM3736-32", "GM3736-32", "GM3736-34", "GM3736-34",
"GM3736-37", "GM3736-37", "GM3736-44", "GM3736-44", "GM3736-51",
"GM3736-51", "GM3736-64", "GM3736-64", "GM3736-70", "GM3736-70",
"GM3736-71", "GM3736-71", "GM3736-72", "GM3736-72", "GM3736-73",
"GM3736-73", "GM3736-75", "GM3736-75", "GM3736-78", "GM3736-78",
"GM3736-82", "GM3736-82", "GM4034-1", "GM4034-1", "GM905-52",
"GM905-52", "GM905-57", "GM905-57", "GM905-60", "GM905-60", "HMC1",
"HMC1", "MAL3", "MAL3", "MEX2", "MEX2", "PAN51", "PAN51", "PAR68",
"PAR68", "PER183", "PER183", "SM1127-8", "SM1127-8", "SM1219-9",
"SM1219-9", "SM2775-4", "SM2775-4", "SM2792-31", "SM2792-31",
"SM2828-28", "SM2828-28", "SM3134-73", "SM3134-73", "SMB2446-2",
"SMB2446-2", "TAI8", "TAI8"), TIPO = c("SEMBRADAS", "GERMINADAS",
"SEMBRADAS", "GERMINADAS", "SEMBRADAS", "GERMINADAS", "SEMBRADAS",
"GERMINADAS", "SEMBRADAS", "GERMINADAS", "SEMBRADAS", "GERMINADAS",
"SEMBRADAS", "GERMINADAS", "SEMBRADAS", "GERMINADAS", "SEMBRADAS",
"GERMINADAS", "SEMBRADAS", "GERMINADAS", "SEMBRADAS", "GERMINADAS",
"SEMBRADAS", "GERMINADAS", "SEMBRADAS", "GERMINADAS", "SEMBRADAS",
"GERMINADAS", "SEMBRADAS", "GERMINADAS", "SEMBRADAS", "GERMINADAS",
"SEMBRADAS", "GERMINADAS", "SEMBRADAS", "GERMINADAS", "SEMBRADAS",
"GERMINADAS", "SEMBRADAS", "GERMINADAS", "SEMBRADAS", "GERMINADAS",
"SEMBRADAS", "GERMINADAS", "SEMBRADAS", "GERMINADAS", "SEMBRADAS",
"GERMINADAS", "SEMBRADAS", "GERMINADAS", "SEMBRADAS", "GERMINADAS",
"SEMBRADAS", "GERMINADAS", "SEMBRADAS", "GERMINADAS", "SEMBRADAS",
"GERMINADAS", "SEMBRADAS", "GERMINADAS", "SEMBRADAS", "GERMINADAS",
"SEMBRADAS", "GERMINADAS", "SEMBRADAS", "GERMINADAS", "SEMBRADAS",
"GERMINADAS", "SEMBRADAS", "GERMINADAS", "SEMBRADAS", "GERMINADAS",
"SEMBRADAS", "GERMINADAS", "SEMBRADAS", "GERMINADAS", "SEMBRADAS",
"GERMINADAS", "SEMBRADAS", "GERMINADAS", "SEMBRADAS", "GERMINADAS",
"SEMBRADAS", "GERMINADAS", "SEMBRADAS", "GERMINADAS", "SEMBRADAS",
"GERMINADAS", "SEMBRADAS", "GERMINADAS", "SEMBRADAS", "GERMINADAS",
"SEMBRADAS", "GERMINADAS", "SEMBRADAS", "GERMINADAS", "SEMBRADAS",
"GERMINADAS", "SEMBRADAS", "GERMINADAS", "SEMBRADAS", "GERMINADAS",
"SEMBRADAS", "GERMINADAS", "SEMBRADAS", "GERMINADAS", "SEMBRADAS",
"GERMINADAS", "SEMBRADAS", "GERMINADAS", "SEMBRADAS", "GERMINADAS",
"SEMBRADAS", "GERMINADAS", "SEMBRADAS", "GERMINADAS", "SEMBRADAS",
"GERMINADAS", "SEMBRADAS", "GERMINADAS"), Valor = c(40, 40, 40,
40, 40, 39, 40, 40, 40, 40, 40, 40, 40, 40, 40, 39, 40, 40, 40,
40, 40, 40, 40, 40, 40, 39, 40, 40, 40, 40, 40, 39, 40, 40, 40,
40, 40, 40, 40, 29, 40, 22, 40, 36, 40, 29, 40, 35, 40, 32, 40,
9, 40, 40, 40, 23, 40, 38, 40, 40, 40, 28, 40, 25, 40, 39, 40,
34, 40, 40, 40, 40, 40, 40, 40, 40, 40, 38, 40, 40, 40, 38, 40,
38, 40, 40, 40, 26, 40, 40, 40, 38, 40, 40, 40, 40, 40, 40, 40,
40, 40, 40, 40, 40, 40, 40, 40, 40, 40, 40, 40, 40, 40, 38, 40,
37, 40, 40, 40, 40)), class = c("tbl_df", "tbl", "data.frame"
), row.names = c(NA, -120L))
# I'm to order acording with Valor.
DF2 |> arrange(-Valor) # this order is not good for me
# plot
DF2 |> arrange(-Valor) |>
slice_head(n=30) |> #View()
ggplot(aes(x=fct_reorder(germplasmName,-Valor),
y=Valor,
fill=TIPO)) +
geom_col()+
scale_fill_manual(values = c('#216A9E','#F79D27')) +
scale_y_continuous(limits = ~ c(0, max(.x) + 4),
expand = c(0, 0))+
facet_wrap(~factor(TIPO,levels=c('SEMBRADAS','GERMINADAS')),
scales = 'free_x')+
geom_text(aes(label=Valor, vjust = -0.15),
size=3.5)+
theme_classic()+
theme(legend.position = 'none',
axis.text.x = element_text(angle = 90, vjust = 0.5, hjust=1))+
labs(title = '202305DVGN6_ciat',
x='Material',
y='Conteo')
Like said, each germplasmName have two factors in TIPO. I need organize according to the maximum Valor but preserving the TIPO factor. This for get the same x axis in the facet_wrap()
The idea is get this order and next make the plot because in this form I can get the two TIPO for each germplasmName acording max Valor
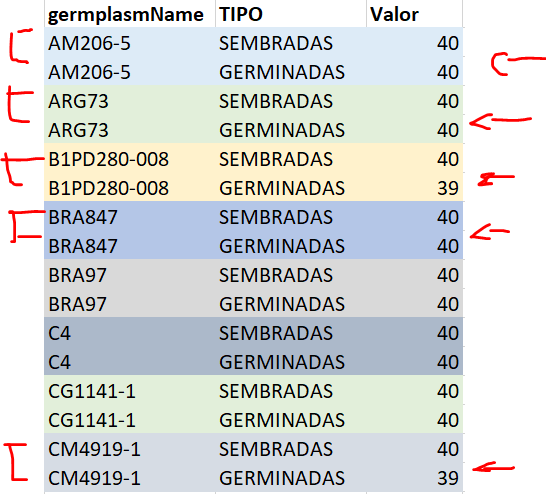
I hope I have explained this topic well.