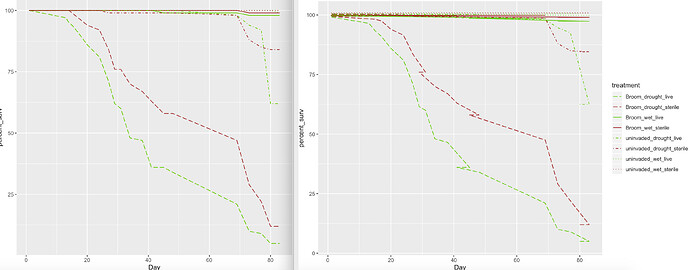
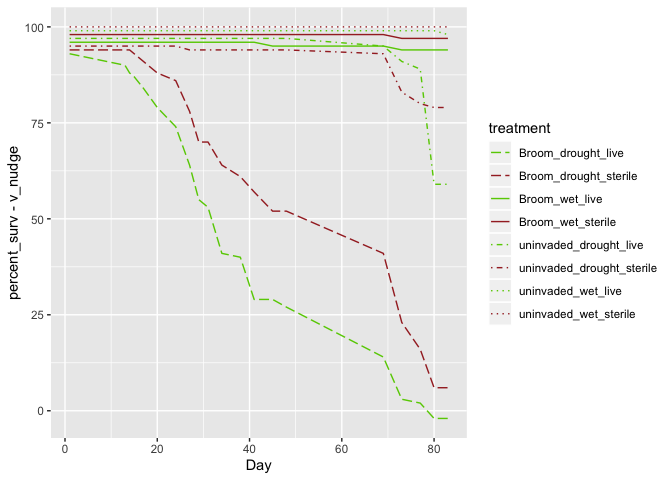
Here's my question:
When I do a vertical dodge on my line graph, I get this weird horizontal shift on the spaces between 2 datapoints that were equal in y value but not in their x value (any time the lines go flat, they do a zig-zag when I apply a vertical dodge to my graph). Can anybody help me solve this issue?
Here's my data and code
dput(cont_surv)
structure(list(Day = c(1L, 13L, 14L, 15L, 17L, 20L, 24L, 27L,
29L, 31L, 34L, 38L, 41L, 45L, 48L, 69L, 73L, 77L, 80L, 83L, 1L,
13L, 14L, 15L, 17L, 20L, 24L, 27L, 29L, 31L, 34L, 38L, 41L, 45L,
48L, 69L, 73L, 77L, 80L, 83L, 1L, 13L, 14L, 15L, 17L, 20L, 24L,
27L, 29L, 31L, 34L, 38L, 41L, 45L, 48L, 69L, 73L, 77L, 80L, 83L,
1L, 13L, 14L, 15L, 17L, 20L, 24L, 27L, 29L, 31L, 34L, 38L, 41L,
45L, 48L, 69L, 73L, 77L, 80L, 83L, 1L, 13L, 14L, 15L, 17L, 20L,
24L, 27L, 29L, 31L, 34L, 38L, 41L, 45L, 48L, 69L, 73L, 77L, 80L,
83L, 1L, 13L, 14L, 15L, 17L, 20L, 24L, 27L, 29L, 31L, 34L, 38L,
41L, 45L, 48L, 69L, 73L, 77L, 80L, 83L, 1L, 13L, 14L, 15L, 17L,
20L, 24L, 27L, 29L, 31L, 34L, 38L, 41L, 45L, 48L, 69L, 73L, 77L,
80L, 83L, 1L, 13L, 14L, 15L, 17L, 20L, 24L, 27L, 29L, 31L, 34L,
38L, 41L, 45L, 48L, 69L, 73L, 77L, 80L, 83L), percent_surv = c(100L,
97L, 95L, 94L, 91L, 86L, 81L, 71L, 62L, 60L, 48L, 47L, 36L, 36L,
34L, 21L, 10L, 9L, 5L, 5L, 100L, 100L, 100L, 99L, 97L, 94L, 92L,
84L, 76L, 76L, 70L, 67L, 63L, 58L, 58L, 47L, 29L, 22L, 12L, 12L,
100L, 100L, 100L, 100L, 100L, 100L, 100L, 100L, 100L, 100L, 100L,
100L, 100L, 99L, 99L, 99L, 98L, 98L, 98L, 98L, 100L, 100L, 100L,
100L, 100L, 100L, 100L, 100L, 100L, 100L, 100L, 100L, 100L, 100L,
100L, 100L, 99L, 99L, 99L, 99L, 100L, 100L, 100L, 100L, 100L,
100L, 100L, 100L, 100L, 100L, 100L, 100L, 100L, 100L, 100L, 98L,
94L, 92L, 62L, 62L, 100L, 100L, 100L, 100L, 100L, 100L, 100L,
99L, 99L, 99L, 99L, 99L, 99L, 99L, 99L, 98L, 88L, 85L, 84L, 84L,
100L, 100L, 100L, 100L, 100L, 100L, 100L, 100L, 100L, 100L, 100L,
100L, 100L, 100L, 100L, 100L, 100L, 100L, 100L, 99L, 100L, 100L,
100L, 100L, 100L, 100L, 100L, 100L, 100L, 100L, 100L, 100L, 100L,
100L, 100L, 100L, 100L, 100L, 100L, 100L), treatment = structure(c(1L,
1L, 1L, 1L, 1L, 1L, 1L, 1L, 1L, 1L, 1L, 1L, 1L, 1L, 1L, 1L, 1L,
1L, 1L, 1L, 2L, 2L, 2L, 2L, 2L, 2L, 2L, 2L, 2L, 2L, 2L, 2L, 2L,
2L, 2L, 2L, 2L, 2L, 2L, 2L, 3L, 3L, 3L, 3L, 3L, 3L, 3L, 3L, 3L,
3L, 3L, 3L, 3L, 3L, 3L, 3L, 3L, 3L, 3L, 3L, 4L, 4L, 4L, 4L, 4L,
4L, 4L, 4L, 4L, 4L, 4L, 4L, 4L, 4L, 4L, 4L, 4L, 4L, 4L, 4L, 5L,
5L, 5L, 5L, 5L, 5L, 5L, 5L, 5L, 5L, 5L, 5L, 5L, 5L, 5L, 5L, 5L,
5L, 5L, 5L, 6L, 6L, 6L, 6L, 6L, 6L, 6L, 6L, 6L, 6L, 6L, 6L, 6L,
6L, 6L, 6L, 6L, 6L, 6L, 6L, 7L, 7L, 7L, 7L, 7L, 7L, 7L, 7L, 7L,
7L, 7L, 7L, 7L, 7L, 7L, 7L, 7L, 7L, 7L, 7L, 8L, 8L, 8L, 8L, 8L,
8L, 8L, 8L, 8L, 8L, 8L, 8L, 8L, 8L, 8L, 8L, 8L, 8L, 8L, 8L), .Label = c("Broom_drought_live",
"Broom_drought_sterile", "Broom_wet_live", "Broom_wet_sterile",
"uninvaded_drought_live", "uninvaded_drought_sterile", "uninvaded_wet_live",
"uninvaded_wet_sterile"), class = "factor")), class = "data.frame", row.names = c(NA,
-160L))
and my code
ggplot(cont_surv,
aes(
x=Day,
y=percent_surv,
group = treatment,
xlab = "Days since drought treatment start",
ylab = "Percent surviving"
)
)+
geom_line(aes(linetype=treatment,
color = treatment
), position=position_dodgev(height=2)
) +
scale_linetype_manual(values= c(5,5,1,1,4,4,3,3)
)+
scale_color_manual(values=c(
"chartreuse3",
"brown",
"chartreuse3",
"brown",
"chartreuse3",
"brown",
"chartreuse3",
"brown"
))+
scale_size_manual(values=c(1,1.5))