Hi everyone!
I am very new to R, and was trying to figure out a way to produce a volcano plot from my data. I was trying to follow the tutorial:
But with my own data. However, I get a couple of errors...
Here is my code:
#edgeR analysis
library(limma)
library(edgeR)
library(readxl)
library(dplyr)
library(ggrepel) # Volcano plot
library(ggplot2) # GO Term plot
# Importing the excel data file containing two healthy datasets to compare
group <- factor(c(rep('HD1', 10), rep('HD2', 10)))
my_data <- read_xlsx('HD1vsHD2.xlsx') %>%
{
DGEList(counts = C[,c(3:31)],
genes = C[, c('GeneID', 'Length')], group = group)
}
# Volcano Plot
pvalue <- 0.05
ggplot(data=my_data, aes(x=log2FoldChange,
y=-log10(pvalue),
col=diffexpressed, label=delabel)) +
geom_point() +
theme_minimal() +
geom_text_repel() +
scale_color_manual(values=c("blue", "black", "red")) +
geom_vline(xintercept=c(-0.6, 0.6), col="red") +
geom_hline(yintercept=-log10(0.05), col="red")
I keep getting:
Error in C[, c(3:28)] : object of type 'closure' is not subsettable
and
Error in FUN(X[[i]], ...) : object 'log2FoldChange' not found
I'm not entirely sure what to do to define the log2FoldChange ![]()
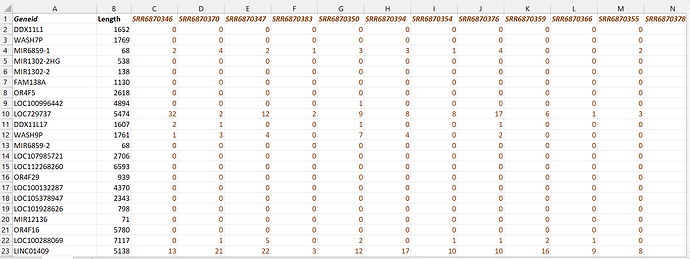
My (excel) data is comprised of RNA gene sequences and the counted lengths in patients:
There are 31 samples total, vertically. I am comparing the first 21 columns vs the last 10 columns of the data in an excel file. I am essentially trying to observe differentiated genes using a volcano plot, but I can't get the syntax to work.
Any help?
Thank you!!